Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
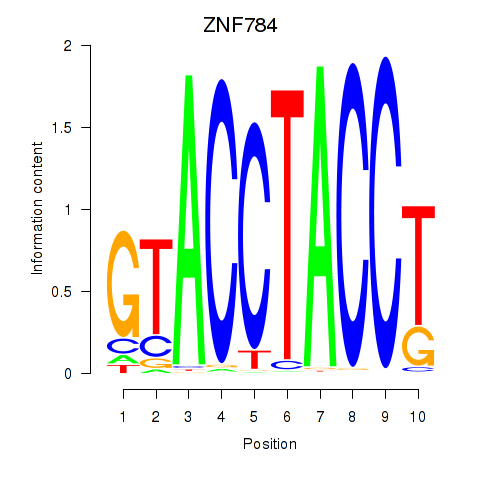
Results for ZNF784
Z-value: 0.90
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.6 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF784 | hg38_v1_chr19_-_55624563_55624570 | -0.23 | 2.1e-01 | Click! |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_26293025 | 2.11 |
ENST00000396376.5
|
SNX10
|
sorting nexin 10 |
| chr17_-_66220630 | 1.80 |
ENST00000585162.1
|
APOH
|
apolipoprotein H |
| chr15_-_101489697 | 1.61 |
ENST00000611967.4
ENST00000615296.4 ENST00000611716.5 ENST00000618548.4 ENST00000619160.4 ENST00000622483.4 ENST00000559417.2 |
PCSK6
|
proprotein convertase subtilisin/kexin type 6 |
| chr19_-_41994217 | 1.47 |
ENST00000648268.1
ENST00000545399.6 |
ATP1A3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr19_-_41994079 | 1.43 |
ENST00000602133.5
|
ATP1A3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr9_-_101442403 | 1.26 |
ENST00000648758.1
|
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr7_-_95434951 | 1.19 |
ENST00000478801.5
ENST00000469926.5 ENST00000493290.5 ENST00000222572.8 ENST00000633531.1 ENST00000490778.5 ENST00000433091.6 |
PON2
|
paraoxonase 2 |
| chr6_-_24489565 | 1.18 |
ENST00000230036.2
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr5_+_161848314 | 1.18 |
ENST00000437025.6
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr12_+_57434778 | 1.17 |
ENST00000309668.3
|
INHBC
|
inhibin subunit beta C |
| chr7_-_95435329 | 1.17 |
ENST00000633192.1
|
PON2
|
paraoxonase 2 |
| chr5_+_161848112 | 1.13 |
ENST00000393943.10
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr10_-_72954790 | 1.10 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB |
| chr2_-_40512361 | 1.03 |
ENST00000403092.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr5_+_161848536 | 1.03 |
ENST00000519621.2
ENST00000636573.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr3_-_42264887 | 0.99 |
ENST00000334681.9
|
CCK
|
cholecystokinin |
| chr2_-_40512423 | 0.99 |
ENST00000402441.5
ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr12_+_50104000 | 0.97 |
ENST00000548814.1
ENST00000301149.8 |
GPD1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr4_+_37453914 | 0.97 |
ENST00000381980.9
ENST00000508175.5 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr16_+_19168207 | 0.91 |
ENST00000355377.7
ENST00000568115.5 |
SYT17
|
synaptotagmin 17 |
| chr9_-_101442372 | 0.91 |
ENST00000648423.1
|
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr9_-_109498251 | 0.90 |
ENST00000374541.4
ENST00000262539.7 |
PTPN3
|
protein tyrosine phosphatase non-receptor type 3 |
| chr3_-_47578832 | 0.89 |
ENST00000264723.9
ENST00000610462.1 |
CSPG5
|
chondroitin sulfate proteoglycan 5 |
| chr6_+_122779707 | 0.88 |
ENST00000368444.8
ENST00000356535.4 |
FABP7
|
fatty acid binding protein 7 |
| chr8_+_79611036 | 0.86 |
ENST00000220876.12
ENST00000518111.5 |
STMN2
|
stathmin 2 |
| chr12_-_95996302 | 0.84 |
ENST00000261208.8
ENST00000538703.5 ENST00000541929.5 |
HAL
|
histidine ammonia-lyase |
| chr2_-_70553638 | 0.83 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr11_-_60183011 | 0.83 |
ENST00000533023.5
ENST00000420732.6 ENST00000528851.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr10_-_125823089 | 0.78 |
ENST00000368774.1
ENST00000368778.7 ENST00000649536.1 |
UROS
|
uroporphyrinogen III synthase |
| chr8_+_22056837 | 0.75 |
ENST00000520174.5
|
DMTN
|
dematin actin binding protein |
| chr3_+_51977833 | 0.75 |
ENST00000637978.1
|
ABHD14A-ACY1
|
ABHD14A-ACY1 readthrough |
| chr10_-_125823221 | 0.74 |
ENST00000420761.5
ENST00000368797.10 |
UROS
|
uroporphyrinogen III synthase |
| chr2_-_40430257 | 0.74 |
ENST00000408028.6
ENST00000332839.8 ENST00000406391.2 ENST00000405901.7 |
SLC8A1
|
solute carrier family 8 member A1 |
| chr5_+_112738331 | 0.73 |
ENST00000512211.6
|
APC
|
APC regulator of WNT signaling pathway |
| chr4_+_157221598 | 0.73 |
ENST00000505888.1
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr15_+_59372685 | 0.72 |
ENST00000558348.5
|
FAM81A
|
family with sequence similarity 81 member A |
| chr11_-_60183191 | 0.72 |
ENST00000412309.6
|
MS4A6A
|
membrane spanning 4-domains A6A |
| chr4_-_82044069 | 0.68 |
ENST00000638048.1
|
RASGEF1B
|
RasGEF domain family member 1B |
| chr12_-_14843517 | 0.68 |
ENST00000228936.6
|
ART4
|
ADP-ribosyltransferase 4 (inactive) (Dombrock blood group) |
| chr1_+_205043204 | 0.67 |
ENST00000331830.7
|
CNTN2
|
contactin 2 |
| chr1_+_205043165 | 0.66 |
ENST00000640428.1
|
CNTN2
|
contactin 2 |
| chr11_-_12008584 | 0.66 |
ENST00000534511.5
ENST00000683431.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr4_-_76898118 | 0.65 |
ENST00000334306.4
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr12_+_128853462 | 0.65 |
ENST00000281703.11
ENST00000442111.6 |
GLT1D1
|
glycosyltransferase 1 domain containing 1 |
| chr10_+_17951906 | 0.64 |
ENST00000377371.3
ENST00000377369.7 |
SLC39A12
|
solute carrier family 39 member 12 |
| chr4_-_82044244 | 0.64 |
ENST00000514050.6
|
RASGEF1B
|
RasGEF domain family member 1B |
| chr19_+_49119531 | 0.64 |
ENST00000334186.9
|
PPFIA3
|
PTPRF interacting protein alpha 3 |
| chr7_+_128937917 | 0.64 |
ENST00000357234.10
ENST00000613821.4 ENST00000477535.5 ENST00000479582.5 ENST00000464557.5 ENST00000402030.6 |
IRF5
|
interferon regulatory factor 5 |
| chr1_+_205042960 | 0.63 |
ENST00000638378.1
|
CNTN2
|
contactin 2 |
| chr17_-_18363504 | 0.63 |
ENST00000583780.1
ENST00000316694.8 ENST00000352886.10 |
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr5_-_134004635 | 0.62 |
ENST00000425992.2
ENST00000395044.7 ENST00000395047.6 |
VDAC1
|
voltage dependent anion channel 1 |
| chr3_+_124584625 | 0.62 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr2_-_73642413 | 0.62 |
ENST00000272425.4
|
NAT8
|
N-acetyltransferase 8 (putative) |
| chr8_+_22056966 | 0.62 |
ENST00000517804.5
|
DMTN
|
dematin actin binding protein |
| chr3_-_39280021 | 0.61 |
ENST00000399220.3
|
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr4_-_152352800 | 0.60 |
ENST00000393956.9
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr2_+_170816868 | 0.60 |
ENST00000358196.8
|
GAD1
|
glutamate decarboxylase 1 |
| chr17_-_18363451 | 0.60 |
ENST00000354098.7
|
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr12_-_49131391 | 0.60 |
ENST00000336023.9
ENST00000550367.1 ENST00000552984.1 ENST00000547476.5 |
TUBA1B
|
tubulin alpha 1b |
| chr3_-_62875029 | 0.59 |
ENST00000490353.2
|
CADPS
|
calcium dependent secretion activator |
| chr12_-_95995920 | 0.59 |
ENST00000552509.5
|
HAL
|
histidine ammonia-lyase |
| chr4_+_157220654 | 0.59 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr17_-_18363346 | 0.58 |
ENST00000582653.1
|
SHMT1
|
serine hydroxymethyltransferase 1 |
| chr19_-_4540028 | 0.57 |
ENST00000306390.7
|
LRG1
|
leucine rich alpha-2-glycoprotein 1 |
| chr3_-_39280432 | 0.56 |
ENST00000542107.5
ENST00000435290.1 |
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr14_-_105864247 | 0.56 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chrX_-_54998530 | 0.56 |
ENST00000545676.5
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_+_44338452 | 0.56 |
ENST00000396078.7
|
TCAIM
|
T cell activation inhibitor, mitochondrial |
| chr2_+_170816562 | 0.56 |
ENST00000625689.2
ENST00000375272.5 |
GAD1
|
glutamate decarboxylase 1 |
| chr10_+_17951885 | 0.55 |
ENST00000377374.8
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr8_+_144466317 | 0.55 |
ENST00000645548.2
ENST00000642354.1 ENST00000301332.3 |
KIFC2
|
kinesin family member C2 |
| chr3_-_121660892 | 0.54 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr7_+_114922561 | 0.54 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr11_+_75151055 | 0.54 |
ENST00000532236.5
|
SLCO2B1
|
solute carrier organic anion transporter family member 2B1 |
| chr10_+_17951825 | 0.53 |
ENST00000539911.5
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr1_+_151060357 | 0.53 |
ENST00000368921.5
|
MLLT11
|
MLLT11 transcription factor 7 cofactor |
| chr13_-_94712505 | 0.52 |
ENST00000376945.4
|
SOX21
|
SRY-box transcription factor 21 |
| chr22_-_26590082 | 0.52 |
ENST00000442495.5
ENST00000440953.5 ENST00000450022.1 ENST00000338754.9 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr4_+_157220691 | 0.50 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr1_+_173868350 | 0.49 |
ENST00000427304.5
ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr5_-_175444132 | 0.49 |
ENST00000393752.3
|
DRD1
|
dopamine receptor D1 |
| chr7_+_8435063 | 0.49 |
ENST00000438764.1
|
NXPH1
|
neurexophilin 1 |
| chr1_-_161238196 | 0.48 |
ENST00000367983.9
ENST00000506209.5 ENST00000367980.6 ENST00000628566.2 |
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr2_-_133568393 | 0.48 |
ENST00000317721.10
ENST00000405974.7 ENST00000409261.6 ENST00000409213.5 |
NCKAP5
|
NCK associated protein 5 |
| chr11_+_73647701 | 0.48 |
ENST00000543524.5
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr6_-_25930678 | 0.47 |
ENST00000377850.8
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr2_+_45651650 | 0.47 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr10_-_46023445 | 0.47 |
ENST00000585132.5
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr1_+_165631199 | 0.47 |
ENST00000367888.8
ENST00000367889.8 ENST00000367885.5 ENST00000367884.6 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr1_+_205504592 | 0.46 |
ENST00000506784.5
ENST00000360066.6 |
CDK18
|
cyclin dependent kinase 18 |
| chr6_-_25930611 | 0.46 |
ENST00000360488.7
|
SLC17A2
|
solute carrier family 17 member 2 |
| chr2_+_172735912 | 0.46 |
ENST00000409036.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr1_+_89821921 | 0.45 |
ENST00000394593.7
|
LRRC8D
|
leucine rich repeat containing 8 VRAC subunit D |
| chr1_+_50109620 | 0.45 |
ENST00000371819.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr1_+_173868076 | 0.45 |
ENST00000367704.5
|
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr1_-_175192769 | 0.45 |
ENST00000423313.6
|
KIAA0040
|
KIAA0040 |
| chr4_+_70397931 | 0.45 |
ENST00000399575.7
|
OPRPN
|
opiorphin prepropeptide |
| chr8_+_30638562 | 0.44 |
ENST00000517349.2
|
SMIM18
|
small integral membrane protein 18 |
| chr4_+_41360759 | 0.44 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_-_52656536 | 0.44 |
ENST00000508499.5
|
USP46
|
ubiquitin specific peptidase 46 |
| chr22_+_20967212 | 0.44 |
ENST00000434714.6
|
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr17_-_10697501 | 0.43 |
ENST00000577427.1
ENST00000255390.10 |
SCO1
|
synthesis of cytochrome C oxidase 1 |
| chr4_-_89836963 | 0.43 |
ENST00000420646.6
ENST00000673718.1 |
SNCA
|
synuclein alpha |
| chr22_+_20967243 | 0.43 |
ENST00000683034.1
ENST00000440238.4 ENST00000405089.5 |
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr7_+_8435187 | 0.43 |
ENST00000429542.1
|
NXPH1
|
neurexophilin 1 |
| chr11_+_125569280 | 0.43 |
ENST00000529812.5
|
EI24
|
EI24 autophagy associated transmembrane protein |
| chr8_+_84182777 | 0.42 |
ENST00000522613.5
|
RALYL
|
RALY RNA binding protein like |
| chr3_+_50674896 | 0.42 |
ENST00000266037.10
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr19_+_18173804 | 0.42 |
ENST00000407280.4
|
IFI30
|
IFI30 lysosomal thiol reductase |
| chr7_-_954666 | 0.41 |
ENST00000265846.10
ENST00000649206.1 |
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr15_-_26939518 | 0.41 |
ENST00000541819.6
|
GABRB3
|
gamma-aminobutyric acid type A receptor subunit beta3 |
| chr4_+_6782631 | 0.40 |
ENST00000508423.1
|
KIAA0232
|
KIAA0232 |
| chr4_-_89837076 | 0.40 |
ENST00000506691.1
|
SNCA
|
synuclein alpha |
| chr1_-_84690255 | 0.40 |
ENST00000603677.1
|
SSX2IP
|
SSX family member 2 interacting protein |
| chr10_+_80408503 | 0.40 |
ENST00000606162.6
|
PRXL2A
|
peroxiredoxin like 2A |
| chr2_-_231926328 | 0.40 |
ENST00000295440.2
ENST00000409852.2 |
NPPC
|
natriuretic peptide C |
| chr2_+_170816905 | 0.40 |
ENST00000344257.9
|
GAD1
|
glutamate decarboxylase 1 |
| chr14_-_105863862 | 0.40 |
ENST00000488476.1
|
IGHJ5
|
immunoglobulin heavy joining 5 |
| chr5_-_74767105 | 0.39 |
ENST00000509430.5
ENST00000296805.8 ENST00000345239.6 ENST00000427854.6 ENST00000506778.1 |
GFM2
|
GTP dependent ribosome recycling factor mitochondrial 2 |
| chr1_-_84690406 | 0.39 |
ENST00000605755.5
ENST00000342203.8 ENST00000437941.6 |
SSX2IP
|
SSX family member 2 interacting protein |
| chr11_-_67504252 | 0.39 |
ENST00000533391.5
ENST00000534749.5 ENST00000532703.5 |
PITPNM1
|
phosphatidylinositol transfer protein membrane associated 1 |
| chr5_+_176549323 | 0.39 |
ENST00000261944.9
|
CDHR2
|
cadherin related family member 2 |
| chr22_+_44180915 | 0.39 |
ENST00000444313.8
ENST00000416291.5 |
PARVG
|
parvin gamma |
| chr3_-_112499457 | 0.39 |
ENST00000334529.10
|
BTLA
|
B and T lymphocyte associated |
| chr17_+_81239310 | 0.39 |
ENST00000431388.3
|
NDUFAF8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr12_+_6951345 | 0.38 |
ENST00000318974.14
ENST00000541698.5 ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chr9_-_96619378 | 0.38 |
ENST00000375240.7
ENST00000463569.5 |
CDC14B
|
cell division cycle 14B |
| chr5_+_69217798 | 0.38 |
ENST00000512880.5
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr5_-_134004921 | 0.38 |
ENST00000265333.8
|
VDAC1
|
voltage dependent anion channel 1 |
| chr16_+_1333631 | 0.38 |
ENST00000397488.6
ENST00000562208.5 ENST00000568887.5 ENST00000426824.8 |
BAIAP3
|
BAI1 associated protein 3 |
| chr12_+_6951271 | 0.38 |
ENST00000456013.5
|
PTPN6
|
protein tyrosine phosphatase non-receptor type 6 |
| chrX_-_155612928 | 0.37 |
ENST00000334398.8
|
TMLHE
|
trimethyllysine hydroxylase, epsilon |
| chr3_+_184276007 | 0.36 |
ENST00000359140.8
ENST00000404464.8 ENST00000357474.9 |
ECE2
|
endothelin converting enzyme 2 |
| chr3_+_16884942 | 0.36 |
ENST00000615277.5
|
PLCL2
|
phospholipase C like 2 |
| chr1_+_50109817 | 0.36 |
ENST00000652353.1
ENST00000371821.6 ENST00000652274.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr7_+_45027634 | 0.35 |
ENST00000381112.7
ENST00000474617.1 |
CCM2
|
CCM2 scaffold protein |
| chr4_-_6200520 | 0.35 |
ENST00000409021.9
ENST00000409371.8 ENST00000282924.9 ENST00000531445.3 |
JAKMIP1
C4orf50
|
janus kinase and microtubule interacting protein 1 chromosome 4 open reading frame 50 |
| chr14_+_22521975 | 0.35 |
ENST00000390515.1
|
TRAJ22
|
T cell receptor alpha joining 22 |
| chr15_+_88803468 | 0.34 |
ENST00000558207.5
|
ACAN
|
aggrecan |
| chr1_+_50109788 | 0.34 |
ENST00000651258.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr11_+_73647645 | 0.34 |
ENST00000545798.5
ENST00000539157.5 ENST00000546251.5 ENST00000535582.5 ENST00000538227.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr4_+_1347197 | 0.34 |
ENST00000389851.10
ENST00000511216.6 |
UVSSA
|
UV stimulated scaffold protein A |
| chr16_-_18433412 | 0.34 |
ENST00000620440.4
|
NOMO2
|
NODAL modulator 2 |
| chr12_-_62191422 | 0.34 |
ENST00000551449.1
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr1_-_161238223 | 0.33 |
ENST00000515452.1
|
NR1I3
|
nuclear receptor subfamily 1 group I member 3 |
| chr21_+_44350126 | 0.33 |
ENST00000300482.9
ENST00000431901.5 |
TRPM2
|
transient receptor potential cation channel subfamily M member 2 |
| chr3_+_54122636 | 0.33 |
ENST00000288197.9
|
CACNA2D3
|
calcium voltage-gated channel auxiliary subunit alpha2delta 3 |
| chrX_-_54357993 | 0.33 |
ENST00000375169.7
ENST00000354646.6 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr1_+_171314171 | 0.33 |
ENST00000367749.4
|
FMO4
|
flavin containing dimethylaniline monoxygenase 4 |
| chr1_-_175192754 | 0.33 |
ENST00000545251.6
|
KIAA0040
|
KIAA0040 |
| chr9_-_76394372 | 0.33 |
ENST00000376736.6
|
RFK
|
riboflavin kinase |
| chr10_+_21534315 | 0.32 |
ENST00000377091.7
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr5_+_129094691 | 0.32 |
ENST00000173527.6
ENST00000514194.5 |
ISOC1
|
isochorismatase domain containing 1 |
| chr1_+_54980610 | 0.32 |
ENST00000371268.4
|
TMEM61
|
transmembrane protein 61 |
| chr15_-_55365231 | 0.32 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr10_+_80408485 | 0.32 |
ENST00000615554.4
ENST00000372185.5 |
PRXL2A
|
peroxiredoxin like 2A |
| chr5_+_112737847 | 0.32 |
ENST00000257430.9
ENST00000508376.6 |
APC
|
APC regulator of WNT signaling pathway |
| chr7_-_103989516 | 0.31 |
ENST00000343529.9
ENST00000424685.3 |
RELN
|
reelin |
| chr3_+_44338119 | 0.31 |
ENST00000383746.7
ENST00000417237.5 |
TCAIM
|
T cell activation inhibitor, mitochondrial |
| chr6_-_31582415 | 0.31 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr15_+_40239857 | 0.31 |
ENST00000260404.8
|
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr16_+_82035245 | 0.31 |
ENST00000199936.9
|
HSD17B2
|
hydroxysteroid 17-beta dehydrogenase 2 |
| chr15_+_40239420 | 0.31 |
ENST00000560346.5
|
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr3_-_112499358 | 0.30 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr21_-_7829587 | 0.30 |
ENST00000623803.1
ENST00000618699.3 |
KCNE1B
|
potassium voltage-gated channel subfamily E regulatory subunit 1B |
| chr12_-_47725518 | 0.30 |
ENST00000545824.2
|
ENDOU
|
endonuclease, poly(U) specific |
| chr4_+_41360597 | 0.30 |
ENST00000509638.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_+_97646034 | 0.30 |
ENST00000258424.3
|
COX5B
|
cytochrome c oxidase subunit 5B |
| chr9_-_96619783 | 0.30 |
ENST00000375241.6
|
CDC14B
|
cell division cycle 14B |
| chr8_+_66712700 | 0.30 |
ENST00000521198.7
|
SGK3
|
serum/glucocorticoid regulated kinase family member 3 |
| chr12_-_54259531 | 0.29 |
ENST00000550411.5
ENST00000439541.6 |
CBX5
|
chromobox 5 |
| chrX_-_50643649 | 0.29 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr9_-_136203183 | 0.29 |
ENST00000371746.9
|
LHX3
|
LIM homeobox 3 |
| chr5_-_74767042 | 0.29 |
ENST00000509097.1
|
GFM2
|
GTP dependent ribosome recycling factor mitochondrial 2 |
| chr9_-_76394132 | 0.29 |
ENST00000490113.1
|
RFK
|
riboflavin kinase |
| chrX_-_155612877 | 0.28 |
ENST00000369439.4
|
TMLHE
|
trimethyllysine hydroxylase, epsilon |
| chrX_+_49162564 | 0.28 |
ENST00000616266.4
|
MAGIX
|
MAGI family member, X-linked |
| chr17_-_28902930 | 0.27 |
ENST00000426464.2
|
DHRS13
|
dehydrogenase/reductase 13 |
| chr12_-_12684490 | 0.27 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr3_+_45026296 | 0.27 |
ENST00000296130.5
|
CLEC3B
|
C-type lectin domain family 3 member B |
| chr18_-_24272179 | 0.27 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr20_-_34951852 | 0.27 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr11_+_826136 | 0.27 |
ENST00000528315.1
ENST00000533803.1 |
CRACR2B
|
calcium release activated channel regulator 2B |
| chr7_+_142349135 | 0.27 |
ENST00000634383.1
|
TRBV6-2
|
T cell receptor beta variable 6-2 |
| chr8_-_23706756 | 0.27 |
ENST00000325017.4
|
NKX2-6
|
NK2 homeobox 6 |
| chr21_-_7829926 | 0.26 |
ENST00000622690.4
|
KCNE1B
|
potassium voltage-gated channel subfamily E regulatory subunit 1B |
| chr8_-_139703093 | 0.26 |
ENST00000650269.1
ENST00000520439.3 |
KCNK9
|
potassium two pore domain channel subfamily K member 9 |
| chr17_-_58417521 | 0.26 |
ENST00000584437.5
ENST00000407977.7 |
RNF43
|
ring finger protein 43 |
| chr12_-_47725483 | 0.26 |
ENST00000422538.8
|
ENDOU
|
endonuclease, poly(U) specific |
| chr14_+_22271921 | 0.26 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr1_-_24964984 | 0.25 |
ENST00000338888.3
ENST00000399916.5 |
RUNX3
|
RUNX family transcription factor 3 |
| chr18_-_55588146 | 0.25 |
ENST00000627784.2
|
TCF4
|
transcription factor 4 |
| chr9_-_112718021 | 0.25 |
ENST00000374234.1
ENST00000374242.9 ENST00000374236.5 |
INIP
|
INTS3 and NABP interacting protein |
| chr1_+_50108856 | 0.25 |
ENST00000650764.1
ENST00000494555.2 ENST00000371824.7 ENST00000371823.8 ENST00000652693.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr12_-_47725558 | 0.25 |
ENST00000229003.7
|
ENDOU
|
endonuclease, poly(U) specific |
| chr3_+_39329700 | 0.24 |
ENST00000326306.5
|
CCR8
|
C-C motif chemokine receptor 8 |
| chr7_+_114922346 | 0.24 |
ENST00000393486.5
|
MDFIC
|
MyoD family inhibitor domain containing |
| chrX_+_155612558 | 0.24 |
ENST00000675360.1
|
SPRY3
|
sprouty RTK signaling antagonist 3 |
| chr7_-_3043838 | 0.23 |
ENST00000356408.3
ENST00000396946.9 |
CARD11
|
caspase recruitment domain family member 11 |
| chr10_-_5185104 | 0.23 |
ENST00000648824.1
ENST00000650030.1 |
AKR1C8P
|
aldo-keto reductase family 1 member C8, pseudogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.6 | 1.8 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.4 | 1.5 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.3 | 1.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.3 | 1.2 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.3 | 1.8 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.3 | 1.2 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.3 | 3.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.2 | 2.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 1.3 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.9 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 2.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 1.4 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 0.6 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.2 | 0.5 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.2 | 0.6 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.8 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.4 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 0.5 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 3.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 1.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 1.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.5 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 1.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.6 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.7 | GO:1902613 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.4 | GO:1900194 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.2 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 1.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 | 2.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 1.0 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 2.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.7 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.7 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 1.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.9 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 1.0 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.0 | 0.1 | GO:1902173 | negative regulation of keratinocyte apoptotic process(GO:1902173) |
| 0.0 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.5 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.3 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1903978 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) regulation of microglial cell activation(GO:1903978) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 1.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:2000606 | regulation of cell proliferation involved in mesonephros development(GO:2000606) negative regulation of cell proliferation involved in mesonephros development(GO:2000607) fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000699) glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000701) regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000702) negative regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation(GO:2000703) regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000733) negative regulation of glial cell-derived neurotrophic factor receptor signaling pathway involved in ureteric bud formation(GO:2000734) |
| 0.0 | 0.1 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.5 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0003290 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.5 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 1.4 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 2.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 2.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.6 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.6 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.1 | 0.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 3.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 1.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 2.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 2.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.5 | 2.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.4 | 1.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.4 | 3.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.4 | 1.8 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 2.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 1.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.2 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.2 | 1.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.0 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.2 | 2.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 2.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.8 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.5 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 1.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 1.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 1.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 1.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 3.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 2.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |