Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
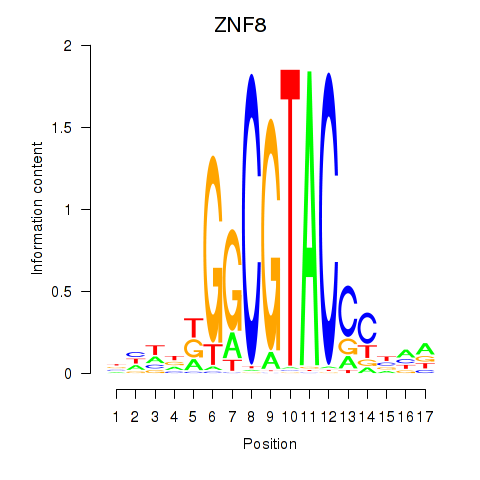
Results for ZNF8
Z-value: 0.55
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000278129.2 | zinc finger protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg38_v1_chr19_+_58278948_58278992 | 0.01 | 9.7e-01 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55140922 | 2.54 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr12_-_46269105 | 2.46 |
ENST00000546893.5
|
SLC38A1
|
solute carrier family 38 member 1 |
| chr12_-_46268989 | 2.31 |
ENST00000549049.5
ENST00000439706.5 ENST00000398637.10 |
SLC38A1
|
solute carrier family 38 member 1 |
| chr6_+_18155399 | 1.48 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr6_+_18155329 | 1.19 |
ENST00000546309.6
|
KDM1B
|
lysine demethylase 1B |
| chr3_+_50617390 | 1.00 |
ENST00000457064.1
|
MAPKAPK3
|
MAPK activated protein kinase 3 |
| chr11_+_13668702 | 0.97 |
ENST00000532701.1
|
FAR1
|
fatty acyl-CoA reductase 1 |
| chr2_+_219491867 | 0.90 |
ENST00000412982.5
|
ENSG00000286143.1
|
novel protein |
| chr6_-_116279837 | 0.83 |
ENST00000368608.4
|
TSPYL1
|
TSPY like 1 |
| chr13_+_52455429 | 0.82 |
ENST00000468284.1
ENST00000378034.7 ENST00000378037.9 ENST00000258607.10 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr2_-_239400949 | 0.80 |
ENST00000345617.7
|
HDAC4
|
histone deacetylase 4 |
| chr2_-_239400991 | 0.75 |
ENST00000543185.6
|
HDAC4
|
histone deacetylase 4 |
| chr9_-_37576365 | 0.72 |
ENST00000432825.7
|
FBXO10
|
F-box protein 10 |
| chr4_-_185425941 | 0.69 |
ENST00000264689.11
ENST00000505357.1 |
UFSP2
|
UFM1 specific peptidase 2 |
| chr11_+_13668653 | 0.67 |
ENST00000354817.8
|
FAR1
|
fatty acyl-CoA reductase 1 |
| chr3_-_57079287 | 0.63 |
ENST00000338458.8
ENST00000468727.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr17_+_35809474 | 0.59 |
ENST00000604879.5
ENST00000603427.5 ENST00000603777.5 ENST00000605844.6 ENST00000604841.5 |
TAF15
|
TATA-box binding protein associated factor 15 |
| chr12_-_122266425 | 0.57 |
ENST00000643696.1
ENST00000267199.9 |
VPS33A
|
VPS33A core subunit of CORVET and HOPS complexes |
| chrX_-_93673558 | 0.56 |
ENST00000475430.2
ENST00000373079.4 |
NAP1L3
|
nucleosome assembly protein 1 like 3 |
| chr3_-_9952337 | 0.54 |
ENST00000411976.2
ENST00000412055.6 |
PRRT3
|
proline rich transmembrane protein 3 |
| chr4_-_52020332 | 0.52 |
ENST00000682860.1
|
LRRC66
|
leucine rich repeat containing 66 |
| chr10_+_988425 | 0.50 |
ENST00000360803.9
|
GTPBP4
|
GTP binding protein 4 |
| chr16_+_21953288 | 0.49 |
ENST00000630839.2
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein 2 |
| chr17_+_50547089 | 0.47 |
ENST00000619622.4
ENST00000356488.8 ENST00000006658.11 |
SPATA20
|
spermatogenesis associated 20 |
| chr9_+_32551670 | 0.47 |
ENST00000450093.3
|
SMIM27
|
small integral membrane protein 27 |
| chr17_-_28842727 | 0.45 |
ENST00000583307.6
ENST00000581229.6 ENST00000582266.6 ENST00000577376.6 ENST00000577682.6 ENST00000581407.6 ENST00000341217.7 ENST00000583522.6 |
FAM222B
|
family with sequence similarity 222 member B |
| chr4_+_169620509 | 0.45 |
ENST00000347613.8
|
CLCN3
|
chloride voltage-gated channel 3 |
| chr7_-_75073844 | 0.42 |
ENST00000614461.4
ENST00000618035.4 |
RCC1L
|
RCC1 like |
| chr9_+_93263948 | 0.42 |
ENST00000448251.5
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr3_-_46717855 | 0.41 |
ENST00000315170.13
|
PRSS50
|
serine protease 50 |
| chr3_-_45958654 | 0.40 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain autophagy adaptor 1 |
| chr20_+_38748448 | 0.39 |
ENST00000243903.6
|
ACTR5
|
actin related protein 5 |
| chr4_+_169620527 | 0.35 |
ENST00000360642.7
ENST00000512813.5 ENST00000513761.6 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr6_-_31795475 | 0.35 |
ENST00000440048.1
|
VARS1
|
valyl-tRNA synthetase 1 |
| chr4_-_76148382 | 0.35 |
ENST00000264883.8
ENST00000514987.5 ENST00000514901.5 |
NUP54
|
nucleoporin 54 |
| chr12_-_122266410 | 0.32 |
ENST00000451053.3
|
VPS33A
|
VPS33A core subunit of CORVET and HOPS complexes |
| chr16_+_21953341 | 0.32 |
ENST00000268379.9
ENST00000561553.5 ENST00000565331.5 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein 2 |
| chr2_-_105329685 | 0.31 |
ENST00000393359.7
|
TGFBRAP1
|
transforming growth factor beta receptor associated protein 1 |
| chr13_-_25287457 | 0.31 |
ENST00000381801.6
|
MTMR6
|
myotubularin related protein 6 |
| chr2_+_171434196 | 0.31 |
ENST00000375255.8
ENST00000539783.5 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr1_-_54801161 | 0.30 |
ENST00000371274.8
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr11_-_46617170 | 0.27 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr7_-_100100716 | 0.26 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr1_-_54801290 | 0.26 |
ENST00000371276.9
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr22_+_29767351 | 0.25 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_-_186375671 | 0.25 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr6_+_44246166 | 0.25 |
ENST00000620073.4
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr6_-_18155053 | 0.24 |
ENST00000309983.5
|
TPMT
|
thiopurine S-methyltransferase |
| chrX_+_1268828 | 0.23 |
ENST00000432318.8
ENST00000381509.8 ENST00000494969.7 ENST00000355805.7 ENST00000355432.8 |
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha |
| chr8_+_38387120 | 0.20 |
ENST00000526356.1
|
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr1_-_100249815 | 0.19 |
ENST00000370131.3
ENST00000681617.1 ENST00000681780.1 ENST00000370132.8 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr1_-_70205531 | 0.19 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40 |
| chr13_+_21176629 | 0.17 |
ENST00000309594.5
|
MRPL57
|
mitochondrial ribosomal protein L57 |
| chr5_+_139341826 | 0.16 |
ENST00000265192.9
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chrX_+_129779930 | 0.16 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr3_+_49199477 | 0.16 |
ENST00000452691.7
ENST00000366429.2 |
IHO1
|
interactor of HORMAD1 1 |
| chr16_-_53503192 | 0.15 |
ENST00000568596.5
ENST00000394657.12 ENST00000570004.5 ENST00000564497.1 ENST00000300245.8 |
AKTIP
|
AKT interacting protein |
| chr9_-_32552553 | 0.15 |
ENST00000379858.1
ENST00000360538.7 ENST00000681750.1 ENST00000680198.1 |
TOPORS
ENSG00000288684.1
|
TOP1 binding arginine/serine rich protein, E3 ubiquitin ligase novel protein |
| chr2_+_169069537 | 0.15 |
ENST00000428522.5
ENST00000450153.1 ENST00000674881.1 ENST00000421653.5 |
DHRS9
|
dehydrogenase/reductase 9 |
| chr19_+_37507129 | 0.14 |
ENST00000586138.5
ENST00000588578.5 ENST00000587986.5 |
ZNF793
|
zinc finger protein 793 |
| chr5_+_103120314 | 0.14 |
ENST00000613674.4
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr13_+_36819214 | 0.13 |
ENST00000255476.3
|
RFXAP
|
regulatory factor X associated protein |
| chr16_+_68539213 | 0.13 |
ENST00000570495.5
ENST00000564323.5 ENST00000562156.5 ENST00000573685.5 ENST00000563169.7 ENST00000611381.4 |
ZFP90
|
ZFP90 zinc finger protein |
| chr17_+_77185210 | 0.11 |
ENST00000431431.6
|
SEC14L1
|
SEC14 like lipid binding 1 |
| chr14_-_100903696 | 0.08 |
ENST00000649591.1
|
RTL1
|
retrotransposon Gag like 1 |
| chr5_-_94111627 | 0.08 |
ENST00000505869.5
ENST00000395965.8 ENST00000509163.5 |
FAM172A
|
family with sequence similarity 172 member A |
| chr5_+_139341875 | 0.07 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr22_-_29766934 | 0.06 |
ENST00000344318.4
|
ZMAT5
|
zinc finger matrin-type 5 |
| chr22_-_38656353 | 0.06 |
ENST00000535113.7
|
FAM227A
|
family with sequence similarity 227 member A |
| chr17_-_58514617 | 0.06 |
ENST00000682306.1
|
MTMR4
|
myotubularin related protein 4 |
| chr9_-_37576336 | 0.06 |
ENST00000541607.1
|
FBXO10
|
F-box protein 10 |
| chr2_+_130182224 | 0.04 |
ENST00000651060.1
ENST00000409255.1 ENST00000281871.11 ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr1_-_235128819 | 0.03 |
ENST00000366607.5
|
TOMM20
|
translocase of outer mitochondrial membrane 20 |
| chr8_+_26291758 | 0.03 |
ENST00000522535.5
ENST00000665949.1 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr5_-_138575359 | 0.03 |
ENST00000297185.9
ENST00000678300.1 ENST00000677425.1 ENST00000677064.1 ENST00000507115.6 |
HSPA9
|
heat shock protein family A (Hsp70) member 9 |
| chr2_+_118096418 | 0.03 |
ENST00000614681.1
|
INSIG2
|
insulin induced gene 2 |
| chr1_-_22965338 | 0.02 |
ENST00000426928.6
|
LACTBL1
|
lactamase beta like 1 |
| chr13_-_21176540 | 0.02 |
ENST00000400018.7
ENST00000314759.6 |
SKA3
|
spindle and kinetochore associated complex subunit 3 |
| chr5_+_103120264 | 0.02 |
ENST00000358359.8
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr2_-_171433950 | 0.01 |
ENST00000375258.9
ENST00000442541.1 ENST00000392599.6 |
METTL8
|
methyltransferase like 8 |
| chrX_+_130401962 | 0.01 |
ENST00000305536.11
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein X-linked 2 |
| chr1_-_228109255 | 0.01 |
ENST00000483159.1
ENST00000366744.5 ENST00000348259.9 ENST00000495434.1 ENST00000295008.8 ENST00000366731.9 ENST00000366746.7 ENST00000366747.7 ENST00000336520.8 ENST00000391867.8 ENST00000457264.5 ENST00000464148.2 ENST00000336300.9 ENST00000430433.5 |
MRPL55
|
mitochondrial ribosomal protein L55 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 2.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.5 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 2.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.6 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.3 | GO:0031453 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) |
| 0.1 | 4.8 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.1 | 0.2 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.1 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.0 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.9 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.2 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.1 | 1.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.6 | GO:0031672 | A band(GO:0031672) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.3 | 1.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 2.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 1.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 4.8 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 0.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 2.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |