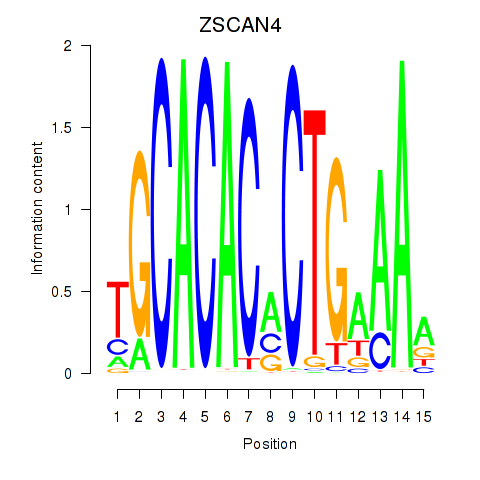
|
chr17_+_2796404
Show fit
|
3.38 |
ENST00000366401.8
ENST00000254695.13
ENST00000542807.1
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chrX_-_20116871
Show fit
|
2.76 |
ENST00000379651.7
ENST00000443379.7
ENST00000379643.10
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr3_+_111674654
Show fit
|
2.53 |
ENST00000636933.1
ENST00000393934.7
ENST00000477665.2
|
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2
|
|
chr3_-_116445458
Show fit
|
2.48 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein
|
|
chr4_-_167234426
Show fit
|
2.31 |
ENST00000541354.5
ENST00000509854.5
ENST00000512681.5
ENST00000357545.9
ENST00000510741.5
ENST00000510403.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr11_-_125592448
Show fit
|
2.26 |
ENST00000648911.1
|
FEZ1
|
fasciculation and elongation protein zeta 1
|
|
chr3_-_116444983
Show fit
|
2.23 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein
|
|
chr7_-_29969232
Show fit
|
2.15 |
ENST00000409497.5
|
SCRN1
|
secernin 1
|
|
chr4_-_167234579
Show fit
|
2.11 |
ENST00000502330.5
ENST00000357154.7
ENST00000421836.6
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr20_+_10218948
Show fit
|
1.95 |
ENST00000430336.1
|
SNAP25
|
synaptosome associated protein 25
|
|
chr12_+_40692413
Show fit
|
1.94 |
ENST00000551295.7
ENST00000547702.5
ENST00000551424.5
|
CNTN1
|
contactin 1
|
|
chr3_-_58587033
Show fit
|
1.80 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr1_-_177164673
Show fit
|
1.70 |
ENST00000424564.2
ENST00000361833.7
|
ASTN1
|
astrotactin 1
|
|
chr11_-_75351609
Show fit
|
1.70 |
ENST00000420843.7
|
ARRB1
|
arrestin beta 1
|
|
chr12_-_16609869
Show fit
|
1.69 |
ENST00000534946.5
|
LMO3
|
LIM domain only 3
|
|
chr8_+_26390362
Show fit
|
1.68 |
ENST00000518611.5
|
BNIP3L
|
BCL2 interacting protein 3 like
|
|
chr11_-_125496122
Show fit
|
1.62 |
ENST00000527534.1
ENST00000278919.8
ENST00000366139.3
|
FEZ1
|
fasciculation and elongation protein zeta 1
|
|
chr1_-_184037695
Show fit
|
1.57 |
ENST00000361927.9
ENST00000649786.1
|
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2
|
|
chrX_-_20116595
Show fit
|
1.54 |
ENST00000452324.3
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr12_+_120650492
Show fit
|
1.54 |
ENST00000351200.6
|
CABP1
|
calcium binding protein 1
|
|
chr19_+_30372364
Show fit
|
1.48 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536
|
|
chr14_+_64704102
Show fit
|
1.47 |
ENST00000555982.5
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3
|
|
chr11_+_65833944
Show fit
|
1.47 |
ENST00000308342.7
|
SNX32
|
sorting nexin 32
|
|
chr1_+_226548747
Show fit
|
1.46 |
ENST00000366788.8
ENST00000366789.5
|
STUM
|
stum, mechanosensory transduction mediator homolog
|
|
chr6_-_152563271
Show fit
|
1.41 |
ENST00000535896.7
ENST00000672122.1
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1
|
|
chr1_+_159171607
Show fit
|
1.41 |
ENST00000368124.8
ENST00000368125.9
ENST00000416746.1
|
CADM3
|
cell adhesion molecule 3
|
|
chr4_-_167234266
Show fit
|
1.34 |
ENST00000511269.5
ENST00000506697.5
ENST00000512042.1
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr19_+_735026
Show fit
|
1.34 |
ENST00000592155.5
ENST00000590161.2
|
PALM
|
paralemmin
|
|
chr7_+_121873317
Show fit
|
1.33 |
ENST00000651863.1
ENST00000652298.1
ENST00000449182.1
|
PTPRZ1
|
protein tyrosine phosphatase receptor type Z1
|
|
chr1_-_161631152
Show fit
|
1.31 |
ENST00000421702.3
ENST00000650385.1
|
FCGR3B
|
Fc fragment of IgG receptor IIIb
|
|
chr16_+_15643267
Show fit
|
1.31 |
ENST00000396355.5
|
NDE1
|
nudE neurodevelopment protein 1
|
|
chr1_-_161631032
Show fit
|
1.30 |
ENST00000534776.1
ENST00000613418.4
ENST00000614870.4
|
FCGR3B
|
Fc fragment of IgG receptor IIIb
|
|
chr8_-_133102477
Show fit
|
1.29 |
ENST00000522119.5
ENST00000523610.5
ENST00000338087.10
ENST00000521302.5
ENST00000519558.5
ENST00000519747.5
ENST00000517648.5
|
SLA
|
Src like adaptor
|
|
chr11_-_75351686
Show fit
|
1.18 |
ENST00000360025.7
|
ARRB1
|
arrestin beta 1
|
|
chr10_+_43437105
Show fit
|
1.16 |
ENST00000456416.6
ENST00000437590.3
ENST00000451167.2
|
ZNF487
|
zinc finger protein 487
|
|
chr19_-_35135180
Show fit
|
1.13 |
ENST00000392225.7
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr1_+_2104710
Show fit
|
1.09 |
ENST00000482686.5
ENST00000486681.5
|
PRKCZ
|
protein kinase C zeta
|
|
chrX_+_1268807
Show fit
|
1.08 |
ENST00000381524.8
ENST00000381529.9
ENST00000412290.6
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr19_-_39320839
Show fit
|
1.08 |
ENST00000248668.5
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1
|
|
chr9_+_122375286
Show fit
|
1.07 |
ENST00000373698.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr10_+_18260715
Show fit
|
1.06 |
ENST00000615785.4
ENST00000617363.4
ENST00000396576.6
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr19_-_9538590
Show fit
|
1.04 |
ENST00000593003.5
|
ZNF426
|
zinc finger protein 426
|
|
chr2_+_10368645
Show fit
|
1.04 |
ENST00000613496.4
|
HPCAL1
|
hippocalcin like 1
|
|
chr3_+_50246888
Show fit
|
1.03 |
ENST00000451956.1
|
GNAI2
|
G protein subunit alpha i2
|
|
chr11_-_18791563
Show fit
|
1.02 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5
|
|
chr6_-_41941728
Show fit
|
1.01 |
ENST00000414200.6
|
CCND3
|
cyclin D3
|
|
chr12_-_16610037
Show fit
|
1.00 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3
|
|
chr19_-_38224215
Show fit
|
0.99 |
ENST00000355526.10
ENST00000420980.7
ENST00000614244.4
|
DPF1
|
double PHD fingers 1
|
|
chrX_+_1268828
Show fit
|
0.99 |
ENST00000432318.8
ENST00000381509.8
ENST00000494969.7
ENST00000355805.7
ENST00000355432.8
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr13_-_114132580
Show fit
|
0.98 |
ENST00000334062.8
|
RASA3
|
RAS p21 protein activator 3
|
|
chr17_+_32021005
Show fit
|
0.93 |
ENST00000327564.11
ENST00000584368.5
ENST00000394713.7
ENST00000341671.11
|
LRRC37B
|
leucine rich repeat containing 37B
|
|
chr2_-_212124901
Show fit
|
0.93 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chr11_+_61950343
Show fit
|
0.92 |
ENST00000378043.9
|
BEST1
|
bestrophin 1
|
|
chr18_+_34593312
Show fit
|
0.92 |
ENST00000591816.6
ENST00000588125.5
ENST00000684610.1
ENST00000683705.1
ENST00000598334.5
ENST00000588684.5
ENST00000554864.7
ENST00000399121.9
ENST00000595022.5
|
DTNA
|
dystrobrevin alpha
|
|
chr6_-_41941795
Show fit
|
0.91 |
ENST00000372991.9
|
CCND3
|
cyclin D3
|
|
chr11_+_61950063
Show fit
|
0.89 |
ENST00000534553.5
|
BEST1
|
bestrophin 1
|
|
chr4_-_167234552
Show fit
|
0.89 |
ENST00000512648.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr3_+_184276007
Show fit
|
0.88 |
ENST00000359140.8
ENST00000404464.8
ENST00000357474.9
|
ECE2
|
endothelin converting enzyme 2
|
|
chr13_+_111114550
Show fit
|
0.88 |
ENST00000317133.9
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7
|
|
chr6_-_41941507
Show fit
|
0.87 |
ENST00000372987.8
|
CCND3
|
cyclin D3
|
|
chr1_-_161549793
Show fit
|
0.83 |
ENST00000443193.6
|
FCGR3A
|
Fc fragment of IgG receptor IIIa
|
|
chr9_+_121286586
Show fit
|
0.83 |
ENST00000545652.6
|
GSN
|
gelsolin
|
|
chr9_+_137139139
Show fit
|
0.81 |
ENST00000371561.8
|
GRIN1
|
glutamate ionotropic receptor NMDA type subunit 1
|
|
chr14_-_93788475
Show fit
|
0.79 |
ENST00000393140.6
|
PRIMA1
|
proline rich membrane anchor 1
|
|
chr1_-_161549892
Show fit
|
0.78 |
ENST00000426740.7
|
FCGR3A
|
Fc fragment of IgG receptor IIIa
|
|
chr11_-_1598294
Show fit
|
0.78 |
ENST00000412090.2
|
KRTAP5-2
|
keratin associated protein 5-2
|
|
chr2_-_223945354
Show fit
|
0.77 |
ENST00000429915.1
|
WDFY1
|
WD repeat and FYVE domain containing 1
|
|
chr8_-_133102623
Show fit
|
0.77 |
ENST00000524345.5
|
SLA
|
Src like adaptor
|
|
chr19_-_40056156
Show fit
|
0.75 |
ENST00000598845.5
ENST00000593605.1
ENST00000221355.10
ENST00000434248.6
|
ZNF780B
|
zinc finger protein 780B
|
|
chr5_+_137889437
Show fit
|
0.75 |
ENST00000508638.5
ENST00000508883.6
ENST00000502810.5
|
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel
|
|
chr6_-_152563349
Show fit
|
0.74 |
ENST00000673281.1
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1
|
|
chr2_-_174598206
Show fit
|
0.73 |
ENST00000392546.6
ENST00000436221.1
|
WIPF1
|
WAS/WASL interacting protein family member 1
|
|
chr11_-_41459592
Show fit
|
0.72 |
ENST00000528697.6
ENST00000530763.5
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr19_-_3801791
Show fit
|
0.72 |
ENST00000590849.1
ENST00000395045.6
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr19_-_42132465
Show fit
|
0.71 |
ENST00000529067.5
ENST00000529952.5
ENST00000342301.8
ENST00000389341.9
|
POU2F2
|
POU class 2 homeobox 2
|
|
chrX_+_1268786
Show fit
|
0.67 |
ENST00000501036.7
ENST00000417535.7
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr2_-_223945322
Show fit
|
0.65 |
ENST00000233055.9
|
WDFY1
|
WD repeat and FYVE domain containing 1
|
|
chr17_-_48722510
Show fit
|
0.64 |
ENST00000290294.5
|
PRAC1
|
PRAC1 small nuclear protein
|
|
chr5_+_153490727
Show fit
|
0.64 |
ENST00000340592.9
|
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1
|
|
chr6_+_29827817
Show fit
|
0.60 |
ENST00000360323.11
ENST00000376818.7
ENST00000376815.3
|
HLA-G
|
major histocompatibility complex, class I, G
|
|
chr2_+_172860038
Show fit
|
0.59 |
ENST00000538974.5
ENST00000540783.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr10_+_18261080
Show fit
|
0.59 |
ENST00000377319.8
ENST00000377331.8
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr12_-_96400365
Show fit
|
0.58 |
ENST00000261211.8
ENST00000543119.6
|
CDK17
|
cyclin dependent kinase 17
|
|
chr13_-_79406175
Show fit
|
0.57 |
ENST00000438724.5
ENST00000622611.4
ENST00000438737.3
|
RBM26
|
RNA binding motif protein 26
|
|
chr8_-_133102643
Show fit
|
0.57 |
ENST00000519341.5
|
SLA
|
Src like adaptor
|
|
chr19_-_42132391
Show fit
|
0.56 |
ENST00000528894.8
ENST00000560804.6
ENST00000560558.5
ENST00000560398.5
ENST00000526816.6
ENST00000625670.2
|
POU2F2
|
POU class 2 homeobox 2
|
|
chrX_-_6227180
Show fit
|
0.56 |
ENST00000381093.6
|
NLGN4X
|
neuroligin 4 X-linked
|
|
chr12_+_6924449
Show fit
|
0.56 |
ENST00000356654.8
|
ATN1
|
atrophin 1
|
|
chr12_-_6631632
Show fit
|
0.55 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr1_-_154608140
Show fit
|
0.54 |
ENST00000529168.2
ENST00000368474.9
ENST00000680305.1
ENST00000648231.2
|
ADAR
|
adenosine deaminase RNA specific
|
|
chr18_+_75210789
Show fit
|
0.53 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr19_+_44259875
Show fit
|
0.53 |
ENST00000592581.5
ENST00000590668.5
ENST00000683810.1
|
ZNF233
|
zinc finger protein 233
|
|
chr8_+_84182777
Show fit
|
0.53 |
ENST00000522613.5
|
RALYL
|
RALY RNA binding protein like
|
|
chrX_-_63351308
Show fit
|
0.53 |
ENST00000374884.3
|
SPIN4
|
spindlin family member 4
|
|
chr1_+_35869750
Show fit
|
0.50 |
ENST00000373206.5
|
AGO1
|
argonaute RISC component 1
|
|
chr8_-_133102874
Show fit
|
0.49 |
ENST00000395352.7
|
SLA
|
Src like adaptor
|
|
chr19_+_47332167
Show fit
|
0.48 |
ENST00000595464.3
|
C5AR2
|
complement component 5a receptor 2
|
|
chr7_+_64045420
Show fit
|
0.48 |
ENST00000647787.1
ENST00000456806.3
|
ENSG00000285544.1
ZNF727
|
novel transcript
zinc finger protein 727
|
|
chr17_+_41265339
Show fit
|
0.47 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6
|
|
chr4_+_15778320
Show fit
|
0.47 |
ENST00000226279.8
|
CD38
|
CD38 molecule
|
|
chr19_+_44212525
Show fit
|
0.47 |
ENST00000391961.6
ENST00000621083.4
ENST00000313040.12
ENST00000586228.5
ENST00000588219.5
ENST00000589707.5
ENST00000588394.5
ENST00000589005.5
|
ZNF227
|
zinc finger protein 227
|
|
chr8_+_38974212
Show fit
|
0.47 |
ENST00000302495.5
|
HTRA4
|
HtrA serine peptidase 4
|
|
chr13_-_52159559
Show fit
|
0.46 |
ENST00000620675.4
ENST00000610828.5
|
NEK3
|
NIMA related kinase 3
|
|
chr15_+_78340344
Show fit
|
0.46 |
ENST00000299529.7
|
CRABP1
|
cellular retinoic acid binding protein 1
|
|
chr4_+_73853290
Show fit
|
0.46 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr17_+_4715438
Show fit
|
0.45 |
ENST00000571206.1
|
ARRB2
|
arrestin beta 2
|
|
chr9_-_89405926
Show fit
|
0.42 |
ENST00000433650.5
|
SEMA4D
|
semaphorin 4D
|
|
chr8_+_43056409
Show fit
|
0.42 |
ENST00000525699.5
ENST00000529687.5
|
FNTA
|
farnesyltransferase, CAAX box, alpha
|
|
chr18_+_34593392
Show fit
|
0.42 |
ENST00000684377.1
|
DTNA
|
dystrobrevin alpha
|
|
chr7_+_142720652
Show fit
|
0.41 |
ENST00000390400.2
|
TRBV28
|
T cell receptor beta variable 28
|
|
chr19_+_56507800
Show fit
|
0.41 |
ENST00000591537.5
ENST00000308031.10
|
ZNF471
|
zinc finger protein 471
|
|
chr3_-_53255990
Show fit
|
0.41 |
ENST00000423525.6
|
TKT
|
transketolase
|
|
chr11_-_33753394
Show fit
|
0.41 |
ENST00000532057.5
ENST00000531080.5
|
FBXO3
|
F-box protein 3
|
|
chr12_-_84911178
Show fit
|
0.40 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15
|
|
chr6_-_15548360
Show fit
|
0.40 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1
|
|
chr8_+_84183534
Show fit
|
0.40 |
ENST00000518566.5
|
RALYL
|
RALY RNA binding protein like
|
|
chr18_+_58864866
Show fit
|
0.40 |
ENST00000588456.5
ENST00000591808.6
ENST00000589481.1
ENST00000591049.1
|
ZNF532
|
zinc finger protein 532
|
|
chr19_-_40090860
Show fit
|
0.39 |
ENST00000599972.1
ENST00000450241.6
ENST00000595687.6
ENST00000340963.9
|
ZNF780A
|
zinc finger protein 780A
|
|
chr9_-_20621835
Show fit
|
0.39 |
ENST00000630269.2
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr12_+_54549586
Show fit
|
0.39 |
ENST00000243052.8
|
PDE1B
|
phosphodiesterase 1B
|
|
chr2_+_240625237
Show fit
|
0.39 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35
|
|
chr11_+_56027654
Show fit
|
0.39 |
ENST00000641320.1
|
OR5AS1
|
olfactory receptor family 5 subfamily AS member 1
|
|
chr7_-_78771265
Show fit
|
0.38 |
ENST00000630991.2
ENST00000629359.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr3_-_53256009
Show fit
|
0.36 |
ENST00000296289.10
ENST00000462138.6
ENST00000423516.5
|
TKT
|
transketolase
|
|
chr1_-_153945225
Show fit
|
0.34 |
ENST00000368646.6
|
DENND4B
|
DENN domain containing 4B
|
|
chr17_-_58328756
Show fit
|
0.34 |
ENST00000268893.10
ENST00000343736.9
|
TSPOAP1
|
TSPO associated protein 1
|
|
chr13_-_52159529
Show fit
|
0.34 |
ENST00000618534.4
ENST00000550841.1
|
NEK3
|
NIMA related kinase 3
|
|
chr12_+_10010627
Show fit
|
0.33 |
ENST00000338896.11
ENST00000396502.5
|
CLEC12B
|
C-type lectin domain family 12 member B
|
|
chr16_-_55875299
Show fit
|
0.33 |
ENST00000520435.2
|
CES5A
|
carboxylesterase 5A
|
|
chr19_+_571274
Show fit
|
0.33 |
ENST00000545507.6
ENST00000346916.9
|
BSG
|
basigin (Ok blood group)
|
|
chr1_-_111488795
Show fit
|
0.32 |
ENST00000472933.2
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3
|
|
chr7_-_78770859
Show fit
|
0.32 |
ENST00000636717.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr5_+_102808057
Show fit
|
0.32 |
ENST00000684043.1
ENST00000682407.1
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr7_-_149126306
Show fit
|
0.32 |
ENST00000483014.1
ENST00000378061.7
|
ZNF425
|
zinc finger protein 425
|
|
chr12_-_10998304
Show fit
|
0.31 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20
|
|
chr19_+_44259903
Show fit
|
0.30 |
ENST00000588489.5
ENST00000391958.6
|
ZNF233
|
zinc finger protein 233
|
|
chr19_-_40090921
Show fit
|
0.30 |
ENST00000595508.1
ENST00000414720.6
ENST00000455521.5
ENST00000595773.5
ENST00000683561.1
|
ENSG00000269749.1
ZNF780A
|
novel transcript
zinc finger protein 780A
|
|
chr6_-_39725387
Show fit
|
0.30 |
ENST00000287152.12
|
KIF6
|
kinesin family member 6
|
|
chr8_+_106580970
Show fit
|
0.29 |
ENST00000517686.5
|
OXR1
|
oxidation resistance 1
|
|
chr3_-_177196402
Show fit
|
0.29 |
ENST00000437738.5
ENST00000424913.5
ENST00000443315.5
|
TBL1XR1
|
TBL1X receptor 1
|
|
chr16_-_55875358
Show fit
|
0.27 |
ENST00000319165.13
ENST00000290567.14
|
CES5A
|
carboxylesterase 5A
|
|
chr1_+_169367934
Show fit
|
0.26 |
ENST00000367807.7
ENST00000329281.6
ENST00000420531.1
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr7_-_78771058
Show fit
|
0.25 |
ENST00000628781.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr4_-_102760976
Show fit
|
0.23 |
ENST00000644159.1
ENST00000642252.1
|
MANBA
|
mannosidase beta
|
|
chr15_+_75258599
Show fit
|
0.23 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family member C
|
|
chr1_-_153312919
Show fit
|
0.23 |
ENST00000683862.1
|
PGLYRP3
|
peptidoglycan recognition protein 3
|
|
chr3_+_130850585
Show fit
|
0.23 |
ENST00000505330.5
ENST00000504381.5
ENST00000507488.6
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1
|
|
chr4_-_102760914
Show fit
|
0.22 |
ENST00000505239.1
ENST00000647097.2
|
MANBA
|
mannosidase beta
|
|
chr6_-_33580229
Show fit
|
0.22 |
ENST00000374467.4
ENST00000442998.6
ENST00000360661.9
|
BAK1
|
BCL2 antagonist/killer 1
|
|
chr13_-_52159844
Show fit
|
0.22 |
ENST00000611833.4
|
NEK3
|
NIMA related kinase 3
|
|
chr15_-_74082550
Show fit
|
0.22 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family member A
|
|
chr15_+_67125707
Show fit
|
0.21 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr11_+_61950370
Show fit
|
0.20 |
ENST00000449131.6
|
BEST1
|
bestrophin 1
|
|
chr7_-_78771108
Show fit
|
0.20 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr11_+_111879482
Show fit
|
0.20 |
ENST00000260276.8
ENST00000530214.5
ENST00000530799.5
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr6_-_145735964
Show fit
|
0.20 |
ENST00000640980.1
ENST00000639423.1
ENST00000611340.5
|
EPM2A
|
EPM2A glucan phosphatase, laforin
|
|
chr20_-_21514046
Show fit
|
0.20 |
ENST00000377142.5
|
NKX2-2
|
NK2 homeobox 2
|
|
chr15_-_37100523
Show fit
|
0.20 |
ENST00000424352.6
ENST00000561208.6
|
MEIS2
|
Meis homeobox 2
|
|
chr22_+_44752552
Show fit
|
0.19 |
ENST00000389774.6
ENST00000356099.11
ENST00000396119.6
ENST00000336963.8
ENST00000412433.5
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr1_+_94418467
Show fit
|
0.19 |
ENST00000315713.5
|
ABCD3
|
ATP binding cassette subfamily D member 3
|
|
chr1_+_61203496
Show fit
|
0.18 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A
|
|
chr11_-_128942849
Show fit
|
0.18 |
ENST00000531399.6
ENST00000458238.6
|
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1
|
|
chr2_+_127535746
Show fit
|
0.18 |
ENST00000428314.5
ENST00000409816.7
|
MYO7B
|
myosin VIIB
|
|
chr18_-_23662868
Show fit
|
0.18 |
ENST00000586087.1
ENST00000592179.6
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr17_-_35432490
Show fit
|
0.18 |
ENST00000304905.10
|
SLFN12
|
schlafen family member 12
|
|
chr19_-_44529525
Show fit
|
0.17 |
ENST00000620096.4
ENST00000614577.4
ENST00000611497.4
ENST00000614924.5
|
CEACAM20
|
CEA cell adhesion molecule 20
|
|
chr1_-_7940825
Show fit
|
0.17 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9
|
|
chr19_-_10420121
Show fit
|
0.17 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37, HSP90 cochaperone
|
|
chr1_+_15438435
Show fit
|
0.16 |
ENST00000375943.6
ENST00000375949.5
|
CTRC
|
chymotrypsin C
|
|
chr19_+_48695952
Show fit
|
0.15 |
ENST00000522966.2
ENST00000425340.3
ENST00000391876.5
|
FUT2
|
fucosyltransferase 2
|
|
chr10_-_48605032
Show fit
|
0.14 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr14_+_52707192
Show fit
|
0.13 |
ENST00000445930.7
ENST00000555339.5
ENST00000556813.1
|
PSMC6
|
proteasome 26S subunit, ATPase 6
|
|
chr6_-_30686624
Show fit
|
0.13 |
ENST00000274853.8
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18
|
|
chr9_-_122228845
Show fit
|
0.12 |
ENST00000394319.8
ENST00000340587.7
|
LHX6
|
LIM homeobox 6
|
|
chr12_+_121804689
Show fit
|
0.12 |
ENST00000619791.1
ENST00000267197.9
|
SETD1B
|
SET domain containing 1B, histone lysine methyltransferase
|
|
chr1_+_8945858
Show fit
|
0.12 |
ENST00000549778.5
ENST00000377443.7
ENST00000480186.7
ENST00000377436.6
ENST00000377442.3
|
CA6
|
carbonic anhydrase 6
|
|
chr14_+_52707178
Show fit
|
0.12 |
ENST00000612399.4
|
PSMC6
|
proteasome 26S subunit, ATPase 6
|
|
chr5_-_83673544
Show fit
|
0.12 |
ENST00000503117.1
ENST00000510978.5
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr1_+_209756101
Show fit
|
0.11 |
ENST00000479796.5
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr16_+_6019663
Show fit
|
0.11 |
ENST00000422070.8
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr11_-_129947461
Show fit
|
0.10 |
ENST00000526082.5
ENST00000304538.10
|
PRDM10
|
PR/SET domain 10
|
|
chr19_+_51127030
Show fit
|
0.10 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig like lectin 9
|
|
chr7_-_99466148
Show fit
|
0.10 |
ENST00000394186.3
ENST00000359832.8
ENST00000449683.5
ENST00000292475.8
ENST00000488775.5
ENST00000523680.1
ENST00000430982.1
ENST00000413834.5
|
ATP5MF
PTCD1
ATP5MF-PTCD1
|
ATP synthase membrane subunit f
pentatricopeptide repeat domain 1
ATP5MF-PTCD1 readthrough
|
|
chr11_+_10455292
Show fit
|
0.09 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr22_+_44031345
Show fit
|
0.09 |
ENST00000444029.5
|
PARVB
|
parvin beta
|
|
chr10_+_38010617
Show fit
|
0.08 |
ENST00000307441.13
ENST00000374618.7
ENST00000628825.2
ENST00000458705.6
ENST00000432900.7
ENST00000469037.2
|
ZNF33A
|
zinc finger protein 33A
|
|
chr1_+_209756032
Show fit
|
0.08 |
ENST00000400959.7
ENST00000367025.8
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr7_-_16420623
Show fit
|
0.08 |
ENST00000676325.1
|
CRPPA
|
CDP-L-ribitol pyrophosphorylase A
|
|
chr7_-_10940123
Show fit
|
0.08 |
ENST00000339600.6
|
NDUFA4
|
NDUFA4 mitochondrial complex associated
|
|
chr8_-_23854796
Show fit
|
0.08 |
ENST00000290271.7
|
STC1
|
stanniocalcin 1
|
|
chr12_+_133037350
Show fit
|
0.08 |
ENST00000438628.6
|
ZNF84
|
zinc finger protein 84
|
|
chr9_-_20622479
Show fit
|
0.07 |
ENST00000380338.9
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr17_-_19745602
Show fit
|
0.06 |
ENST00000444455.5
ENST00000439102.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1
|
|
chr3_-_195811857
Show fit
|
0.06 |
ENST00000349607.8
ENST00000346145.8
|
MUC4
|
mucin 4, cell surface associated
|
|
chrX_+_17375194
Show fit
|
0.06 |
ENST00000676302.1
|
NHS
|
NHS actin remodeling regulator
|
|
chr12_-_47771029
Show fit
|
0.06 |
ENST00000549151.5
ENST00000548919.5
|
RAPGEF3
|
Rap guanine nucleotide exchange factor 3
|
|
chr1_-_25430147
Show fit
|
0.05 |
ENST00000349320.7
|
RHCE
|
Rh blood group CcEe antigens
|
|
chr1_+_169368175
Show fit
|
0.05 |
ENST00000367808.8
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr2_+_149118169
Show fit
|
0.05 |
ENST00000450639.5
|
LYPD6B
|
LY6/PLAUR domain containing 6B
|
|
chr10_+_113679159
Show fit
|
0.04 |
ENST00000621345.4
ENST00000429617.5
ENST00000369331.8
|
CASP7
|
caspase 7
|