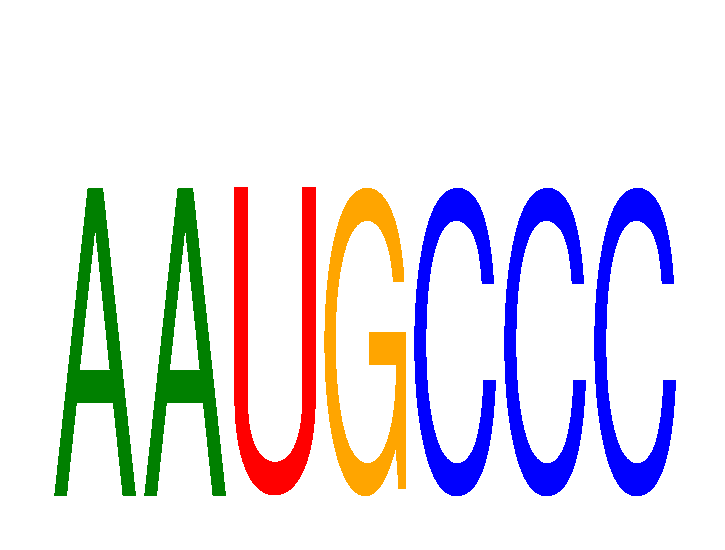Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.80
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-365a-3p
|
MIMAT0000710 |
|
hsa-miR-365b-3p
|
MIMAT0022834 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_134318097 | 1.34 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_+_134525649 | 1.20 |
ENST00000282605.8
ENST00000681547.2 ENST00000361895.6 ENST00000402835.5 |
JADE2
|
jade family PHD finger 2 |
| chr5_-_91383310 | 1.06 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3 |
| chr10_+_99659430 | 1.05 |
ENST00000370489.5
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr9_+_113876282 | 0.94 |
ENST00000374126.9
ENST00000615615.4 ENST00000288466.11 |
ZNF618
|
zinc finger protein 618 |
| chr19_+_46347063 | 0.85 |
ENST00000012443.9
ENST00000391919.1 |
PPP5C
|
protein phosphatase 5 catalytic subunit |
| chr7_-_139777986 | 0.79 |
ENST00000406875.8
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr11_+_2444986 | 0.77 |
ENST00000155840.12
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr2_-_157874976 | 0.75 |
ENST00000682025.1
ENST00000683487.1 ENST00000682300.1 ENST00000683441.1 ENST00000684595.1 ENST00000683426.1 ENST00000683820.1 ENST00000263640.7 |
ACVR1
|
activin A receptor type 1 |
| chr3_+_114056728 | 0.65 |
ENST00000485050.5
ENST00000281273.8 ENST00000479882.5 ENST00000493014.1 |
QTRT2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr11_+_128694052 | 0.64 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_-_51151238 | 0.61 |
ENST00000566102.1
|
SALL1
|
spalt like transcription factor 1 |
| chr12_+_67648737 | 0.61 |
ENST00000344096.4
ENST00000393555.3 |
DYRK2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr15_+_51829644 | 0.60 |
ENST00000308580.12
|
TMOD3
|
tropomodulin 3 |
| chr18_-_63319987 | 0.59 |
ENST00000398117.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr16_-_79600698 | 0.58 |
ENST00000393350.1
|
MAF
|
MAF bZIP transcription factor |
| chr22_+_36913620 | 0.58 |
ENST00000403662.8
ENST00000262825.9 |
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr14_-_99272184 | 0.56 |
ENST00000357195.8
|
BCL11B
|
BAF chromatin remodeling complex subunit BCL11B |
| chr10_+_87863595 | 0.56 |
ENST00000371953.8
|
PTEN
|
phosphatase and tensin homolog |
| chr8_-_56211257 | 0.53 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr17_+_4710622 | 0.51 |
ENST00000574954.5
ENST00000269260.7 ENST00000346341.6 ENST00000572457.5 ENST00000381488.10 ENST00000412477.7 ENST00000571428.5 ENST00000575877.5 |
ARRB2
|
arrestin beta 2 |
| chr15_+_74782069 | 0.51 |
ENST00000220003.14
ENST00000439220.6 |
CSK
|
C-terminal Src kinase |
| chrX_-_3713593 | 0.49 |
ENST00000262848.6
|
PRKX
|
protein kinase X-linked |
| chr6_+_106098933 | 0.49 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr11_-_57515686 | 0.48 |
ENST00000533263.1
ENST00000278426.8 |
SLC43A1
|
solute carrier family 43 member 1 |
| chr20_-_40689228 | 0.47 |
ENST00000373313.3
|
MAFB
|
MAF bZIP transcription factor B |
| chr11_+_47980538 | 0.44 |
ENST00000613246.4
ENST00000418331.7 ENST00000615445.4 ENST00000440289.6 |
PTPRJ
|
protein tyrosine phosphatase receptor type J |
| chr10_-_32347109 | 0.44 |
ENST00000469059.2
ENST00000319778.11 |
EPC1
|
enhancer of polycomb homolog 1 |
| chr4_+_38664189 | 0.43 |
ENST00000514033.1
ENST00000261438.10 |
KLF3
|
Kruppel like factor 3 |
| chr7_-_27165517 | 0.42 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr3_+_172040554 | 0.40 |
ENST00000336824.8
ENST00000423424.5 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr21_+_36699100 | 0.40 |
ENST00000290399.11
|
SIM2
|
SIM bHLH transcription factor 2 |
| chr15_-_58749569 | 0.40 |
ENST00000402627.5
ENST00000559053.1 ENST00000260408.8 ENST00000561288.1 ENST00000461408.2 ENST00000439637.5 ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chrX_-_130110679 | 0.39 |
ENST00000335997.11
|
ELF4
|
E74 like ETS transcription factor 4 |
| chr2_+_191678122 | 0.38 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr12_-_42144823 | 0.38 |
ENST00000398675.8
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chrX_-_110318062 | 0.36 |
ENST00000372059.6
ENST00000262844.10 |
AMMECR1
|
AMMECR nuclear protein 1 |
| chr5_-_100903252 | 0.36 |
ENST00000231461.10
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr16_-_67806513 | 0.36 |
ENST00000317506.8
ENST00000448631.6 ENST00000602677.5 ENST00000630626.2 |
RANBP10
|
RAN binding protein 10 |
| chr5_-_39074377 | 0.35 |
ENST00000514735.1
ENST00000357387.8 ENST00000296782.9 |
RICTOR
|
RPTOR independent companion of MTOR complex 2 |
| chr1_+_2228310 | 0.34 |
ENST00000378536.5
|
SKI
|
SKI proto-oncogene |
| chr5_-_176537361 | 0.34 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44 |
| chr3_+_43286512 | 0.33 |
ENST00000454177.5
ENST00000429705.6 ENST00000296088.12 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr3_-_113746218 | 0.33 |
ENST00000497255.1
ENST00000240922.8 ENST00000478020.1 ENST00000493900.5 |
NAA50
|
N-alpha-acetyltransferase 50, NatE catalytic subunit |
| chr11_-_68841909 | 0.32 |
ENST00000265641.10
ENST00000376618.6 |
CPT1A
|
carnitine palmitoyltransferase 1A |
| chr17_+_70169516 | 0.31 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr17_+_57256514 | 0.31 |
ENST00000284073.7
ENST00000674964.1 |
MSI2
|
musashi RNA binding protein 2 |
| chr11_+_64306227 | 0.31 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chr13_-_45851730 | 0.31 |
ENST00000400405.4
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr17_-_29294141 | 0.31 |
ENST00000225388.9
|
NUFIP2
|
nuclear FMR1 interacting protein 2 |
| chr13_+_113584683 | 0.30 |
ENST00000375370.10
|
TFDP1
|
transcription factor Dp-1 |
| chr2_+_134120169 | 0.30 |
ENST00000409645.5
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr12_-_31326111 | 0.30 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr1_+_35807974 | 0.30 |
ENST00000373210.4
|
AGO4
|
argonaute RISC component 4 |
| chr12_-_89526253 | 0.30 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr11_-_128522264 | 0.30 |
ENST00000531611.5
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr2_-_207769889 | 0.29 |
ENST00000295417.4
|
FZD5
|
frizzled class receptor 5 |
| chr3_-_33440343 | 0.29 |
ENST00000283629.8
|
UBP1
|
upstream binding protein 1 |
| chr14_+_55051639 | 0.28 |
ENST00000395468.9
ENST00000622254.1 |
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1 like |
| chr3_-_47781837 | 0.28 |
ENST00000254480.10
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| chr12_-_89524734 | 0.28 |
ENST00000529983.3
|
GALNT4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr12_-_49060742 | 0.28 |
ENST00000301067.12
ENST00000683543.1 |
KMT2D
|
lysine methyltransferase 2D |
| chr10_+_1049476 | 0.28 |
ENST00000358220.5
|
WDR37
|
WD repeat domain 37 |
| chr14_-_36520222 | 0.27 |
ENST00000354822.7
|
NKX2-1
|
NK2 homeobox 1 |
| chr16_+_53054973 | 0.26 |
ENST00000447540.6
ENST00000615216.4 ENST00000566029.5 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_-_155911340 | 0.26 |
ENST00000368323.8
|
RIT1
|
Ras like without CAAX 1 |
| chr1_+_3069160 | 0.26 |
ENST00000511072.5
|
PRDM16
|
PR/SET domain 16 |
| chr11_+_63839086 | 0.26 |
ENST00000350490.11
ENST00000402010.8 |
MARK2
|
microtubule affinity regulating kinase 2 |
| chr2_+_60881515 | 0.25 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit |
| chr7_+_4682252 | 0.25 |
ENST00000328914.5
|
FOXK1
|
forkhead box K1 |
| chr5_-_168579319 | 0.25 |
ENST00000522176.1
ENST00000239231.7 |
PANK3
|
pantothenate kinase 3 |
| chr1_-_38873322 | 0.24 |
ENST00000397572.5
ENST00000494695.4 |
MYCBP
|
MYC binding protein |
| chr2_-_152717966 | 0.24 |
ENST00000410080.8
|
PRPF40A
|
pre-mRNA processing factor 40 homolog A |
| chr12_+_63844663 | 0.24 |
ENST00000355086.8
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr4_-_148444674 | 0.24 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr10_+_93993897 | 0.24 |
ENST00000371380.8
|
PLCE1
|
phospholipase C epsilon 1 |
| chr2_-_64653906 | 0.23 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr1_+_203305510 | 0.23 |
ENST00000290551.5
|
BTG2
|
BTG anti-proliferation factor 2 |
| chr6_+_16129077 | 0.23 |
ENST00000356840.8
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr9_+_111896804 | 0.23 |
ENST00000374279.4
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr17_-_16215520 | 0.22 |
ENST00000582357.5
ENST00000436828.5 ENST00000268712.8 ENST00000411510.5 ENST00000395857.7 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr1_-_91021455 | 0.22 |
ENST00000347275.9
ENST00000370440.5 |
ZNF644
|
zinc finger protein 644 |
| chr1_-_156082412 | 0.22 |
ENST00000532414.3
|
MEX3A
|
mex-3 RNA binding family member A |
| chr5_-_43313403 | 0.22 |
ENST00000325110.11
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chrX_+_78104229 | 0.22 |
ENST00000373316.5
|
PGK1
|
phosphoglycerate kinase 1 |
| chr7_-_27180013 | 0.21 |
ENST00000470747.4
|
ENSG00000257184.3
|
HOXA10-HOXA9 readthrough |
| chr10_-_96586975 | 0.21 |
ENST00000371142.9
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr1_-_37859583 | 0.21 |
ENST00000373036.5
|
MTF1
|
metal regulatory transcription factor 1 |
| chr18_-_61892997 | 0.20 |
ENST00000312828.4
|
RNF152
|
ring finger protein 152 |
| chr15_+_96330691 | 0.20 |
ENST00000394166.8
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr17_-_42121330 | 0.20 |
ENST00000225916.10
|
KAT2A
|
lysine acetyltransferase 2A |
| chr12_+_459925 | 0.20 |
ENST00000266383.10
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyltransferase 3 |
| chr11_-_18634332 | 0.20 |
ENST00000336349.6
|
SPTY2D1
|
SPT2 chromatin protein domain containing 1 |
| chr15_+_68054308 | 0.20 |
ENST00000249636.11
|
PIAS1
|
protein inhibitor of activated STAT 1 |
| chrX_-_84188148 | 0.20 |
ENST00000262752.5
|
RPS6KA6
|
ribosomal protein S6 kinase A6 |
| chr10_-_118754956 | 0.19 |
ENST00000369151.8
|
CACUL1
|
CDK2 associated cullin domain 1 |
| chr19_-_49072699 | 0.19 |
ENST00000221444.2
|
KCNA7
|
potassium voltage-gated channel subfamily A member 7 |
| chr9_+_127786018 | 0.19 |
ENST00000373264.5
|
CDK9
|
cyclin dependent kinase 9 |
| chr8_+_123768431 | 0.19 |
ENST00000334705.12
ENST00000521166.5 |
FAM91A1
|
family with sequence similarity 91 member A1 |
| chr17_+_16381083 | 0.18 |
ENST00000535788.1
ENST00000302182.8 |
UBB
|
ubiquitin B |
| chr1_-_84997079 | 0.18 |
ENST00000284027.5
ENST00000370608.8 |
MCOLN2
|
mucolipin TRP cation channel 2 |
| chr7_-_141702151 | 0.18 |
ENST00000536163.6
|
DENND11
|
DENN domain containing 11 |
| chr7_+_138460238 | 0.18 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr6_-_111483190 | 0.18 |
ENST00000368802.8
|
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr3_-_125375249 | 0.17 |
ENST00000484491.5
ENST00000492394.5 ENST00000471196.1 ENST00000468369.5 ENST00000485866.5 ENST00000360647.9 |
ZNF148
|
zinc finger protein 148 |
| chr2_+_46698909 | 0.16 |
ENST00000650611.1
ENST00000306503.5 |
LINC01118
SOCS5
|
long intergenic non-protein coding RNA 1118 suppressor of cytokine signaling 5 |
| chr2_-_43226594 | 0.16 |
ENST00000282388.4
|
ZFP36L2
|
ZFP36 ring finger protein like 2 |
| chr13_-_52450590 | 0.16 |
ENST00000378060.9
|
VPS36
|
vacuolar protein sorting 36 homolog |
| chr1_-_21783189 | 0.16 |
ENST00000400301.5
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr12_+_65824475 | 0.16 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2 |
| chr10_+_100745711 | 0.16 |
ENST00000370296.6
ENST00000428433.5 |
PAX2
|
paired box 2 |
| chr15_+_41231219 | 0.15 |
ENST00000334660.10
ENST00000560397.5 |
CHP1
|
calcineurin like EF-hand protein 1 |
| chr3_+_128153422 | 0.15 |
ENST00000254730.11
ENST00000483457.1 |
EEFSEC
|
eukaryotic elongation factor, selenocysteine-tRNA specific |
| chr4_-_73258785 | 0.15 |
ENST00000639793.1
ENST00000358602.9 ENST00000330838.10 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr8_+_22089140 | 0.15 |
ENST00000289921.8
|
FAM160B2
|
family with sequence similarity 160 member B2 |
| chr5_+_112976757 | 0.15 |
ENST00000389063.3
|
DCP2
|
decapping mRNA 2 |
| chr1_+_28736927 | 0.15 |
ENST00000373812.8
ENST00000541996.5 ENST00000496288.5 |
YTHDF2
|
YTH N6-methyladenosine RNA binding protein 2 |
| chr1_-_23531206 | 0.15 |
ENST00000361729.3
|
E2F2
|
E2F transcription factor 2 |
| chr3_+_14947568 | 0.14 |
ENST00000413118.5
ENST00000425241.5 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr8_-_38382146 | 0.14 |
ENST00000534155.1
ENST00000433384.6 ENST00000317025.13 ENST00000316985.7 |
NSD3
|
nuclear receptor binding SET domain protein 3 |
| chr1_-_28088564 | 0.14 |
ENST00000373863.3
ENST00000373871.8 ENST00000540618.5 ENST00000436342.6 |
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr9_-_6008469 | 0.14 |
ENST00000399933.8
|
KIAA2026
|
KIAA2026 |
| chr10_-_102114935 | 0.14 |
ENST00000361198.9
|
LDB1
|
LIM domain binding 1 |
| chr10_+_135067 | 0.14 |
ENST00000381591.5
|
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr7_+_98106852 | 0.13 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr12_+_50085325 | 0.13 |
ENST00000551966.5
ENST00000550477.5 ENST00000394963.9 |
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr10_+_22321056 | 0.13 |
ENST00000376663.8
|
BMI1
|
BMI1 proto-oncogene, polycomb ring finger |
| chr19_+_49591170 | 0.13 |
ENST00000418929.7
|
PRR12
|
proline rich 12 |
| chr1_-_153615858 | 0.13 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14 |
| chr16_+_70523782 | 0.13 |
ENST00000566095.6
ENST00000577085.1 ENST00000302516.10 ENST00000567654.1 |
SF3B3
|
splicing factor 3b subunit 3 |
| chr22_+_20917398 | 0.13 |
ENST00000354336.8
|
CRKL
|
CRK like proto-oncogene, adaptor protein |
| chr10_-_7787946 | 0.13 |
ENST00000379562.9
|
KIN
|
Kin17 DNA and RNA binding protein |
| chr2_-_239400949 | 0.12 |
ENST00000345617.7
|
HDAC4
|
histone deacetylase 4 |
| chr4_-_11428868 | 0.12 |
ENST00000002596.6
|
HS3ST1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr5_+_75337211 | 0.12 |
ENST00000287936.9
ENST00000343975.9 |
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr2_-_189784356 | 0.12 |
ENST00000392350.7
|
ORMDL1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr17_+_60600178 | 0.11 |
ENST00000629650.2
ENST00000305921.8 |
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent 1D |
| chr8_+_58411333 | 0.11 |
ENST00000399598.7
|
UBXN2B
|
UBX domain protein 2B |
| chr4_-_102827494 | 0.11 |
ENST00000453744.7
ENST00000321805.11 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr5_-_90529511 | 0.11 |
ENST00000500869.6
ENST00000315948.11 ENST00000509384.5 |
LYSMD3
|
LysM domain containing 3 |
| chr14_-_21437235 | 0.11 |
ENST00000430710.8
ENST00000646340.1 ENST00000646063.1 |
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr12_+_55743110 | 0.11 |
ENST00000257868.10
|
GDF11
|
growth differentiation factor 11 |
| chr17_-_58007217 | 0.10 |
ENST00000258962.5
ENST00000582730.6 ENST00000584773.5 ENST00000585096.1 |
SRSF1
|
serine and arginine rich splicing factor 1 |
| chr22_+_31944500 | 0.10 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta |
| chr12_-_75511594 | 0.10 |
ENST00000438169.6
ENST00000229214.9 |
KRR1
|
KRR1 small subunit processome component homolog |
| chr9_+_128683645 | 0.09 |
ENST00000372692.8
|
SET
|
SET nuclear proto-oncogene |
| chr17_-_28951285 | 0.09 |
ENST00000577226.5
|
PHF12
|
PHD finger protein 12 |
| chr9_-_134068012 | 0.09 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3 |
| chr1_+_161314372 | 0.08 |
ENST00000342751.8
ENST00000367975.7 ENST00000432287.6 ENST00000392169.6 ENST00000513009.5 |
SDHC
|
succinate dehydrogenase complex subunit C |
| chr1_-_155241220 | 0.08 |
ENST00000368373.8
ENST00000427500.7 |
GBA
|
glucosylceramidase beta |
| chr21_-_44873671 | 0.08 |
ENST00000330938.8
ENST00000397887.7 |
PTTG1IP
|
PTTG1 interacting protein |
| chr3_-_48847797 | 0.08 |
ENST00000454963.5
ENST00000296446.12 ENST00000419216.1 ENST00000265563.13 |
PRKAR2A
|
protein kinase cAMP-dependent type II regulatory subunit alpha |
| chr3_-_71725365 | 0.08 |
ENST00000425534.8
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr15_-_64703199 | 0.08 |
ENST00000559753.1
ENST00000560258.6 ENST00000559912.2 ENST00000326005.10 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chr8_+_127409026 | 0.08 |
ENST00000465342.4
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr5_+_122845488 | 0.08 |
ENST00000513881.5
|
SNX24
|
sorting nexin 24 |
| chr5_+_149730260 | 0.07 |
ENST00000360453.8
ENST00000394320.7 ENST00000309241.10 |
PPARGC1B
|
PPARG coactivator 1 beta |
| chr11_+_66268575 | 0.07 |
ENST00000527397.1
ENST00000311481.11 |
RAB1B
|
RAB1B, member RAS oncogene family |
| chr7_+_139359846 | 0.06 |
ENST00000619796.4
ENST00000354926.9 |
LUC7L2
|
LUC7 like 2, pre-mRNA splicing factor |
| chr7_+_33904911 | 0.06 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator |
| chr17_+_30477362 | 0.06 |
ENST00000225724.9
ENST00000451249.7 ENST00000467337.6 ENST00000581721.5 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chrX_-_75156272 | 0.06 |
ENST00000620875.5
ENST00000669573.1 ENST00000339447.8 ENST00000645829.3 ENST00000529949.5 ENST00000373394.8 ENST00000253577.9 ENST00000644766.1 ENST00000534524.5 |
ABCB7
|
ATP binding cassette subfamily B member 7 |
| chr12_-_48788995 | 0.06 |
ENST00000550422.5
ENST00000357869.8 |
ADCY6
|
adenylate cyclase 6 |
| chrX_-_154751017 | 0.06 |
ENST00000369575.7
ENST00000369568.8 ENST00000424127.3 |
GAB3
|
GRB2 associated binding protein 3 |
| chr1_-_35193135 | 0.06 |
ENST00000357214.6
|
SFPQ
|
splicing factor proline and glutamine rich |
| chrX_-_71068311 | 0.05 |
ENST00000374274.8
|
SNX12
|
sorting nexin 12 |
| chr1_+_35931076 | 0.05 |
ENST00000397828.3
ENST00000373191.9 |
AGO3
|
argonaute RISC catalytic component 3 |
| chr16_+_67029133 | 0.05 |
ENST00000290858.11
ENST00000412916.7 ENST00000564034.6 |
CBFB
|
core-binding factor subunit beta |
| chr12_+_69470349 | 0.04 |
ENST00000547219.5
ENST00000550316.5 ENST00000548154.5 ENST00000547414.5 ENST00000549921.6 ENST00000550389.5 ENST00000550937.5 ENST00000549092.5 ENST00000550169.5 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr7_-_32891744 | 0.04 |
ENST00000304056.9
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr17_-_49995210 | 0.04 |
ENST00000434704.2
|
DLX3
|
distal-less homeobox 3 |
| chr12_-_66678934 | 0.04 |
ENST00000545666.5
ENST00000398016.7 ENST00000359742.9 ENST00000538211.5 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr17_-_80035862 | 0.03 |
ENST00000310924.7
|
TBC1D16
|
TBC1 domain family member 16 |
| chr21_-_26845402 | 0.02 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr6_-_87702221 | 0.02 |
ENST00000257787.6
|
AKIRIN2
|
akirin 2 |
| chr1_-_205750167 | 0.02 |
ENST00000367142.5
|
NUCKS1
|
nuclear casein kinase and cyclin dependent kinase substrate 1 |
| chr12_+_74537787 | 0.02 |
ENST00000519948.4
|
ATXN7L3B
|
ataxin 7 like 3B |
| chr8_-_4994696 | 0.02 |
ENST00000400186.7
ENST00000602723.5 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr5_-_59893718 | 0.02 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D |
| chr14_-_23057530 | 0.02 |
ENST00000397359.7
ENST00000610348.1 |
CDH24
|
cadherin 24 |
| chr6_+_30326835 | 0.01 |
ENST00000440271.5
ENST00000376656.8 ENST00000396551.7 ENST00000428728.5 ENST00000396548.5 ENST00000428404.5 |
TRIM39
|
tripartite motif containing 39 |
| chr6_+_34889228 | 0.01 |
ENST00000360359.5
ENST00000649117.1 ENST00000650178.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr22_-_41621014 | 0.01 |
ENST00000263256.7
|
DESI1
|
desumoylating isopeptidase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060957 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.2 | 1.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.2 | 0.5 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.6 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.5 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.6 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.5 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.6 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.1 | 1.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.7 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 1.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 1.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.4 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.2 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 | 0.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.1 | 0.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.8 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.1 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.4 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 1.0 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.2 | GO:0031049 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.2 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.5 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.4 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.5 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.4 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.1 | 0.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.6 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.3 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.1 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |