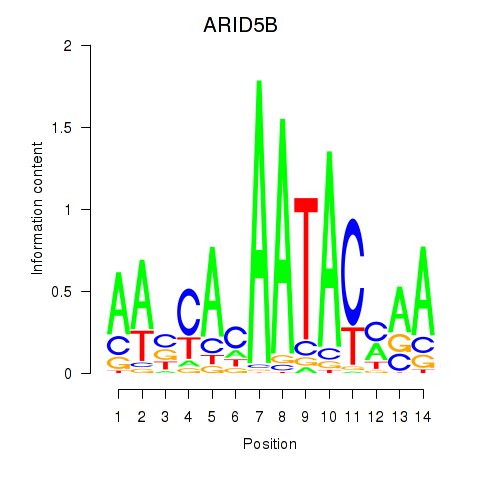
|
chr4_-_70666961
Show fit
|
2.33 |
ENST00000510437.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM
|
|
chrX_+_103330206
Show fit
|
2.29 |
ENST00000372666.1
ENST00000332431.5
|
TCEAL7
|
transcription elongation factor A like 7
|
|
chr4_-_70666884
Show fit
|
2.00 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM
|
|
chr12_-_8662808
Show fit
|
1.96 |
ENST00000359478.7
ENST00000396549.6
|
MFAP5
|
microfibril associated protein 5
|
|
chr1_-_168729187
Show fit
|
1.87 |
ENST00000367817.4
|
DPT
|
dermatopontin
|
|
chr12_-_8662073
Show fit
|
1.82 |
ENST00000535411.5
ENST00000540087.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr2_-_89222461
Show fit
|
1.70 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28
|
|
chr12_-_8662619
Show fit
|
1.57 |
ENST00000544889.1
ENST00000543369.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr12_+_78864768
Show fit
|
1.54 |
ENST00000261205.9
ENST00000457153.6
|
SYT1
|
synaptotagmin 1
|
|
chr2_+_89959979
Show fit
|
1.52 |
ENST00000453166.2
|
IGKV2D-28
|
immunoglobulin kappa variable 2D-28
|
|
chr4_-_70666492
Show fit
|
1.50 |
ENST00000254801.9
ENST00000391614.7
|
JCHAIN
|
joining chain of multimeric IgA and IgM
|
|
chr3_-_98522869
Show fit
|
1.49 |
ENST00000502288.5
ENST00000512147.5
ENST00000341181.11
ENST00000510541.5
ENST00000503621.5
ENST00000511081.5
ENST00000507874.5
ENST00000502299.5
ENST00000508659.5
ENST00000510545.5
ENST00000511667.5
ENST00000394185.6
|
CLDND1
|
claudin domain containing 1
|
|
chr4_-_158173004
Show fit
|
1.46 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B
|
|
chr4_-_158173042
Show fit
|
1.45 |
ENST00000592057.1
ENST00000393807.9
|
GASK1B
|
golgi associated kinase 1B
|
|
chr2_-_89027700
Show fit
|
1.43 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11
|
|
chr3_-_98522754
Show fit
|
1.41 |
ENST00000513287.5
ENST00000514537.5
ENST00000508071.1
ENST00000507944.5
|
CLDND1
|
claudin domain containing 1
|
|
chr3_-_98522514
Show fit
|
1.38 |
ENST00000503004.5
ENST00000506575.1
ENST00000513452.5
ENST00000515620.5
|
CLDND1
|
claudin domain containing 1
|
|
chr17_+_3475959
Show fit
|
1.37 |
ENST00000263080.3
|
ASPA
|
aspartoacylase
|
|
chr6_+_126340107
Show fit
|
1.34 |
ENST00000368328.5
ENST00000368326.5
ENST00000368325.5
|
CENPW
|
centromere protein W
|
|
chr12_-_8662703
Show fit
|
1.32 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr20_+_46021665
Show fit
|
1.31 |
ENST00000454036.6
ENST00000628272.1
ENST00000626701.1
ENST00000413737.2
|
SLC12A5
|
solute carrier family 12 member 5
|
|
chr22_+_22098683
Show fit
|
1.30 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61
|
|
chr12_-_14885845
Show fit
|
1.25 |
ENST00000539261.6
ENST00000228938.5
|
MGP
|
matrix Gla protein
|
|
chr17_-_68955332
Show fit
|
1.13 |
ENST00000269080.6
ENST00000615593.4
ENST00000586539.6
ENST00000430352.6
|
ABCA8
|
ATP binding cassette subfamily A member 8
|
|
chr15_-_79971164
Show fit
|
1.12 |
ENST00000335661.6
ENST00000267953.4
ENST00000677151.1
|
BCL2A1
|
BCL2 related protein A1
|
|
chr12_+_78863962
Show fit
|
1.12 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1
|
|
chr7_+_36410510
Show fit
|
1.10 |
ENST00000428612.5
|
ANLN
|
anillin actin binding protein
|
|
chr4_+_77605807
Show fit
|
1.02 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13
|
|
chr1_+_186828941
Show fit
|
0.98 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA
|
|
chr3_-_98517096
Show fit
|
0.97 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1
|
|
chr8_+_142700095
Show fit
|
0.95 |
ENST00000292430.10
ENST00000518841.5
ENST00000519387.1
|
LY6K
|
lymphocyte antigen 6 family member K
|
|
chr4_+_76074701
Show fit
|
0.94 |
ENST00000355810.9
ENST00000349321.7
|
ART3
|
ADP-ribosyltransferase 3 (inactive)
|
|
chr9_+_111525148
Show fit
|
0.91 |
ENST00000358151.8
ENST00000309235.6
ENST00000355824.7
ENST00000374374.3
|
ZNF483
|
zinc finger protein 483
|
|
chr12_-_8650529
Show fit
|
0.90 |
ENST00000543467.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr10_-_96358989
Show fit
|
0.88 |
ENST00000371172.8
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein
|
|
chr17_+_7407838
Show fit
|
0.85 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2
|
|
chr3_-_151384741
Show fit
|
0.85 |
ENST00000302632.4
|
P2RY12
|
purinergic receptor P2Y12
|
|
chr22_-_21735776
Show fit
|
0.84 |
ENST00000339468.8
|
YPEL1
|
yippee like 1
|
|
chr15_+_51377247
Show fit
|
0.84 |
ENST00000396399.6
|
GLDN
|
gliomedin
|
|
chr11_+_1099730
Show fit
|
0.83 |
ENST00000674892.1
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming
|
|
chr16_-_3372666
Show fit
|
0.83 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4
|
|
chr1_+_103617427
Show fit
|
0.83 |
ENST00000423678.2
ENST00000414303.7
|
AMY2A
|
amylase alpha 2A
|
|
chr1_+_86993009
Show fit
|
0.81 |
ENST00000370548.3
|
ENSG00000267561.2
|
novel protein
|
|
chr12_-_6700788
Show fit
|
0.80 |
ENST00000320591.9
ENST00000534837.6
|
PIANP
|
PILR alpha associated neural protein
|
|
chr11_-_111923722
Show fit
|
0.78 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B
|
|
chr7_+_143382996
Show fit
|
0.76 |
ENST00000446634.1
|
ZYX
|
zyxin
|
|
chr3_+_63443076
Show fit
|
0.74 |
ENST00000295894.9
|
SYNPR
|
synaptoporin
|
|
chr2_+_69013414
Show fit
|
0.74 |
ENST00000681816.1
ENST00000482235.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chr11_+_60327250
Show fit
|
0.73 |
ENST00000684409.1
|
MS4A6E
|
membrane spanning 4-domains A6E
|
|
chr5_+_161850597
Show fit
|
0.73 |
ENST00000634335.1
ENST00000635880.1
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1
|
|
chr7_+_154052373
Show fit
|
0.69 |
ENST00000377770.8
ENST00000406326.5
|
DPP6
|
dipeptidyl peptidase like 6
|
|
chr12_-_91146195
Show fit
|
0.68 |
ENST00000548218.1
|
DCN
|
decorin
|
|
chr20_+_36091409
Show fit
|
0.66 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr18_+_34593312
Show fit
|
0.65 |
ENST00000591816.6
ENST00000588125.5
ENST00000684610.1
ENST00000683705.1
ENST00000598334.5
ENST00000588684.5
ENST00000554864.7
ENST00000399121.9
ENST00000595022.5
|
DTNA
|
dystrobrevin alpha
|
|
chr3_-_79767987
Show fit
|
0.63 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1
|
|
chr22_-_21735744
Show fit
|
0.62 |
ENST00000403503.1
|
YPEL1
|
yippee like 1
|
|
chr17_+_62370218
Show fit
|
0.61 |
ENST00000450662.7
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr15_+_51377410
Show fit
|
0.60 |
ENST00000612989.1
|
GLDN
|
gliomedin
|
|
chr2_-_127675459
Show fit
|
0.60 |
ENST00000355119.9
|
LIMS2
|
LIM zinc finger domain containing 2
|
|
chr22_+_19479826
Show fit
|
0.59 |
ENST00000437685.6
ENST00000263201.7
ENST00000404724.7
|
CDC45
|
cell division cycle 45
|
|
chr12_+_55931148
Show fit
|
0.57 |
ENST00000549629.5
ENST00000555218.5
ENST00000331886.10
|
DGKA
|
diacylglycerol kinase alpha
|
|
chr1_+_151766655
Show fit
|
0.56 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr18_+_34976928
Show fit
|
0.56 |
ENST00000591734.5
ENST00000413393.5
ENST00000589180.5
ENST00000587359.5
|
MAPRE2
|
microtubule associated protein RP/EB family member 2
|
|
chr5_+_175861628
Show fit
|
0.55 |
ENST00000509837.5
|
CPLX2
|
complexin 2
|
|
chr10_-_126521439
Show fit
|
0.55 |
ENST00000284694.11
ENST00000432642.5
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr3_+_99638475
Show fit
|
0.53 |
ENST00000452013.5
ENST00000261037.7
ENST00000652472.1
ENST00000273342.8
ENST00000621757.1
|
COL8A1
|
collagen type VIII alpha 1 chain
|
|
chr4_-_175891691
Show fit
|
0.52 |
ENST00000507540.1
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_84181630
Show fit
|
0.51 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta
|
|
chr2_+_241558716
Show fit
|
0.51 |
ENST00000318407.5
|
BOK
|
BCL2 family apoptosis regulator BOK
|
|
chr1_+_43389874
Show fit
|
0.50 |
ENST00000372450.8
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr7_-_105269007
Show fit
|
0.48 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2
|
|
chr14_+_32329256
Show fit
|
0.48 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6
|
|
chr1_-_168137430
Show fit
|
0.47 |
ENST00000546300.5
ENST00000271357.9
ENST00000367835.1
ENST00000537209.5
|
GPR161
|
G protein-coupled receptor 161
|
|
chr17_-_78736846
Show fit
|
0.47 |
ENST00000589768.5
|
CYTH1
|
cytohesin 1
|
|
chr2_+_69013379
Show fit
|
0.46 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1
|
|
chrX_-_102155790
Show fit
|
0.46 |
ENST00000543160.5
ENST00000333643.4
|
BEX5
|
brain expressed X-linked 5
|
|
chr5_+_119333151
Show fit
|
0.44 |
ENST00000513374.1
|
TNFAIP8
|
TNF alpha induced protein 8
|
|
chr1_+_159005953
Show fit
|
0.44 |
ENST00000426592.6
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr22_+_19479457
Show fit
|
0.44 |
ENST00000407835.6
ENST00000455750.6
|
CDC45
|
cell division cycle 45
|
|
chr8_+_142700465
Show fit
|
0.44 |
ENST00000522591.1
|
LY6K
|
lymphocyte antigen 6 family member K
|
|
chr20_-_9839026
Show fit
|
0.43 |
ENST00000378429.3
ENST00000353224.10
|
PAK5
|
p21 (RAC1) activated kinase 5
|
|
chr1_+_77779618
Show fit
|
0.43 |
ENST00000370791.7
ENST00000443751.3
ENST00000645756.1
ENST00000643390.1
ENST00000642959.1
|
MIGA1
|
mitoguardin 1
|
|
chr2_+_205682491
Show fit
|
0.42 |
ENST00000360409.7
ENST00000450507.5
ENST00000357785.10
ENST00000417189.5
|
NRP2
|
neuropilin 2
|
|
chr5_-_88883701
Show fit
|
0.41 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr12_+_55931447
Show fit
|
0.40 |
ENST00000549368.5
|
DGKA
|
diacylglycerol kinase alpha
|
|
chr1_+_43389889
Show fit
|
0.39 |
ENST00000562955.2
ENST00000634258.3
|
SZT2
|
SZT2 subunit of KICSTOR complex
|
|
chr8_+_73991345
Show fit
|
0.39 |
ENST00000284818.7
ENST00000518893.1
|
LY96
|
lymphocyte antigen 96
|
|
chr7_-_37449174
Show fit
|
0.39 |
ENST00000445322.1
ENST00000448602.5
|
ELMO1
|
engulfment and cell motility 1
|
|
chr3_+_156037693
Show fit
|
0.38 |
ENST00000472028.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1
|
|
chr3_-_74521140
Show fit
|
0.38 |
ENST00000263665.6
|
CNTN3
|
contactin 3
|
|
chr7_-_55552741
Show fit
|
0.37 |
ENST00000418904.5
|
VOPP1
|
VOPP1 WW domain binding protein
|
|
chr19_+_17215332
Show fit
|
0.36 |
ENST00000263897.10
|
USE1
|
unconventional SNARE in the ER 1
|
|
chr14_+_32329341
Show fit
|
0.35 |
ENST00000557354.5
ENST00000557102.1
ENST00000557272.1
|
AKAP6
|
A-kinase anchoring protein 6
|
|
chr1_+_61203496
Show fit
|
0.34 |
ENST00000663597.1
|
NFIA
|
nuclear factor I A
|
|
chr6_+_68638540
Show fit
|
0.34 |
ENST00000546190.5
|
ADGRB3
|
adhesion G protein-coupled receptor B3
|
|
chr3_+_158110363
Show fit
|
0.34 |
ENST00000683137.1
|
RSRC1
|
arginine and serine rich coiled-coil 1
|
|
chr16_+_30199255
Show fit
|
0.34 |
ENST00000338971.10
|
SULT1A3
|
sulfotransferase family 1A member 3
|
|
chr10_-_73591330
Show fit
|
0.33 |
ENST00000451492.5
ENST00000681793.1
ENST00000680396.1
ENST00000413442.5
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr3_+_158110052
Show fit
|
0.32 |
ENST00000295930.7
ENST00000471994.5
ENST00000482822.3
ENST00000476899.6
ENST00000683899.1
ENST00000684604.1
ENST00000682164.1
ENST00000464171.5
ENST00000611884.5
ENST00000312179.10
ENST00000475278.6
|
RSRC1
|
arginine and serine rich coiled-coil 1
|
|
chr10_-_73433550
Show fit
|
0.32 |
ENST00000299432.7
|
MSS51
|
MSS51 mitochondrial translational activator
|
|
chr13_-_41132728
Show fit
|
0.31 |
ENST00000379485.2
|
KBTBD6
|
kelch repeat and BTB domain containing 6
|
|
chr15_+_51377315
Show fit
|
0.31 |
ENST00000558426.5
|
GLDN
|
gliomedin
|
|
chr3_-_113441487
Show fit
|
0.30 |
ENST00000393845.9
ENST00000295868.6
|
CFAP44
|
cilia and flagella associated protein 44
|
|
chr13_+_51861963
Show fit
|
0.30 |
ENST00000242819.7
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr5_-_179620933
Show fit
|
0.30 |
ENST00000521173.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1
|
|
chr6_-_30113086
Show fit
|
0.30 |
ENST00000376734.4
|
TRIM31
|
tripartite motif containing 31
|
|
chr11_-_104898670
Show fit
|
0.29 |
ENST00000422698.6
|
CASP12
|
caspase 12 (gene/pseudogene)
|
|
chr6_+_32844789
Show fit
|
0.29 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9
|
|
chr8_+_66021545
Show fit
|
0.29 |
ENST00000276570.10
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta
|
|
chr1_-_85276467
Show fit
|
0.28 |
ENST00000648566.1
|
BCL10
|
BCL10 immune signaling adaptor
|
|
chrX_-_52797421
Show fit
|
0.28 |
ENST00000375511.4
|
SPANXN5
|
SPANX family member N5
|
|
chr8_-_115492221
Show fit
|
0.28 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1
|
|
chrX_+_129779930
Show fit
|
0.27 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr14_+_22096017
Show fit
|
0.27 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1
|
|
chr4_+_69931066
Show fit
|
0.27 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1
|
|
chr6_+_167291329
Show fit
|
0.27 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A
|
|
chr12_+_92702843
Show fit
|
0.26 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7
|
|
chr15_+_57599411
Show fit
|
0.26 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein
|
|
chr12_+_50842920
Show fit
|
0.26 |
ENST00000551456.5
ENST00000398458.4
|
TMPRSS12
|
transmembrane serine protease 12
|
|
chr17_-_50707855
Show fit
|
0.24 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr5_+_126462339
Show fit
|
0.24 |
ENST00000502348.5
|
GRAMD2B
|
GRAM domain containing 2B
|
|
chr12_+_120496075
Show fit
|
0.23 |
ENST00000548214.1
ENST00000392508.2
|
DYNLL1
|
dynein light chain LC8-type 1
|
|
chr7_+_121076570
Show fit
|
0.23 |
ENST00000443817.1
|
CPED1
|
cadherin like and PC-esterase domain containing 1
|
|
chr9_+_74615582
Show fit
|
0.23 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B
|
|
chr2_-_2324642
Show fit
|
0.22 |
ENST00000650485.1
ENST00000649207.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr1_-_113871665
Show fit
|
0.22 |
ENST00000528414.5
ENST00000460620.5
ENST00000359785.10
ENST00000420377.6
ENST00000525799.1
ENST00000538253.5
|
PTPN22
|
protein tyrosine phosphatase non-receptor type 22
|
|
chr4_-_86101922
Show fit
|
0.22 |
ENST00000472236.5
ENST00000641881.1
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr7_+_31687208
Show fit
|
0.22 |
ENST00000409146.3
ENST00000342032.8
|
PPP1R17
|
protein phosphatase 1 regulatory subunit 17
|
|
chr8_-_100309368
Show fit
|
0.22 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chr8_-_100309926
Show fit
|
0.22 |
ENST00000341084.7
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase
|
|
chrX_+_141002583
Show fit
|
0.22 |
ENST00000449283.2
|
SPANXB1
|
SPANX family member B1
|
|
chr12_+_120496101
Show fit
|
0.21 |
ENST00000550178.1
ENST00000550845.5
ENST00000549989.1
ENST00000552870.1
ENST00000242577.11
|
DYNLL1
|
dynein light chain LC8-type 1
|
|
chr13_-_29586788
Show fit
|
0.20 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 member 1
|
|
chr7_-_26995237
Show fit
|
0.20 |
ENST00000432747.1
|
SKAP2
|
src kinase associated phosphoprotein 2
|
|
chr14_+_21990357
Show fit
|
0.19 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16
|
|
chr19_-_45076504
Show fit
|
0.19 |
ENST00000622376.1
|
ZNF296
|
zinc finger protein 296
|
|
chr5_+_156326735
Show fit
|
0.19 |
ENST00000435422.7
|
SGCD
|
sarcoglycan delta
|
|
chr1_+_43147890
Show fit
|
0.19 |
ENST00000335282.5
ENST00000409396.5
|
FAM183A
|
family with sequence similarity 183 member A
|
|
chr9_+_4985227
Show fit
|
0.19 |
ENST00000381652.4
|
JAK2
|
Janus kinase 2
|
|
chrX_-_143517473
Show fit
|
0.19 |
ENST00000370503.2
|
SPANXN3
|
SPANX family member N3
|
|
chr3_+_196867856
Show fit
|
0.19 |
ENST00000445299.6
ENST00000323460.10
ENST00000419026.5
|
SENP5
|
SUMO specific peptidase 5
|
|
chr2_+_149118169
Show fit
|
0.18 |
ENST00000450639.5
|
LYPD6B
|
LY6/PLAUR domain containing 6B
|
|
chr5_+_137889437
Show fit
|
0.18 |
ENST00000508638.5
ENST00000508883.6
ENST00000502810.5
|
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel
|
|
chr1_+_77779763
Show fit
|
0.18 |
ENST00000646892.1
ENST00000645526.1
|
MIGA1
|
mitoguardin 1
|
|
chr1_+_171512032
Show fit
|
0.18 |
ENST00000426496.6
|
PRRC2C
|
proline rich coiled-coil 2C
|
|
chr6_+_24774925
Show fit
|
0.18 |
ENST00000356509.7
ENST00000230056.8
|
GMNN
|
geminin DNA replication inhibitor
|
|
chr18_-_55635948
Show fit
|
0.18 |
ENST00000565124.4
ENST00000398339.5
|
TCF4
|
transcription factor 4
|
|
chr2_-_74392025
Show fit
|
0.17 |
ENST00000440727.1
ENST00000409240.5
|
DCTN1
|
dynactin subunit 1
|
|
chr17_-_10549612
Show fit
|
0.17 |
ENST00000532183.6
ENST00000397183.6
ENST00000420805.1
|
MYH2
|
myosin heavy chain 2
|
|
chr17_+_39200275
Show fit
|
0.17 |
ENST00000225430.9
|
RPL19
|
ribosomal protein L19
|
|
chr17_+_39200302
Show fit
|
0.17 |
ENST00000579374.5
|
RPL19
|
ribosomal protein L19
|
|
chr17_+_39200334
Show fit
|
0.16 |
ENST00000579260.5
ENST00000582193.5
|
RPL19
|
ribosomal protein L19
|
|
chr12_+_55931070
Show fit
|
0.16 |
ENST00000555090.5
|
DGKA
|
diacylglycerol kinase alpha
|
|
chr6_-_15548360
Show fit
|
0.16 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1
|
|
chr4_+_146175702
Show fit
|
0.16 |
ENST00000296581.11
ENST00000649747.1
ENST00000502781.5
|
LSM6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated
|
|
chr6_+_89483255
Show fit
|
0.16 |
ENST00000520458.5
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr3_-_150703943
Show fit
|
0.16 |
ENST00000491361.5
|
ERICH6
|
glutamate rich 6
|
|
chr15_+_24823625
Show fit
|
0.15 |
ENST00000400100.5
ENST00000645002.1
ENST00000642807.1
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr17_+_39200507
Show fit
|
0.15 |
ENST00000678573.1
|
RPL19
|
ribosomal protein L19
|
|
chr9_-_32552553
Show fit
|
0.15 |
ENST00000379858.1
ENST00000360538.7
ENST00000681750.1
ENST00000680198.1
|
TOPORS
ENSG00000288684.1
|
TOP1 binding arginine/serine rich protein, E3 ubiquitin ligase
novel protein
|
|
chr5_+_132658165
Show fit
|
0.15 |
ENST00000617259.2
ENST00000304506.7
|
IL13
|
interleukin 13
|
|
chr3_-_52679713
Show fit
|
0.14 |
ENST00000296302.11
ENST00000356770.8
ENST00000337303.8
ENST00000409057.5
ENST00000410007.5
ENST00000409114.7
ENST00000409767.5
ENST00000423351.5
|
PBRM1
|
polybromo 1
|
|
chr3_-_132684685
Show fit
|
0.14 |
ENST00000512094.5
ENST00000632629.1
|
NPHP3
NPHP3-ACAD11
|
nephrocystin 3
NPHP3-ACAD11 readthrough (NMD candidate)
|
|
chr1_-_86383078
Show fit
|
0.14 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like
|
|
chr11_+_102112445
Show fit
|
0.14 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator
|
|
chr9_+_113505256
Show fit
|
0.14 |
ENST00000374136.5
|
RGS3
|
regulator of G protein signaling 3
|
|
chr10_+_124461800
Show fit
|
0.14 |
ENST00000368842.10
ENST00000392757.8
ENST00000368839.1
|
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase
|
|
chr3_+_68004231
Show fit
|
0.13 |
ENST00000478136.6
|
TAFA1
|
TAFA chemokine like family member 1
|
|
chr6_-_137173644
Show fit
|
0.13 |
ENST00000296980.7
ENST00000349184.8
ENST00000339602.3
|
IL22RA2
|
interleukin 22 receptor subunit alpha 2
|
|
chr19_-_45076465
Show fit
|
0.13 |
ENST00000303809.7
|
ZNF296
|
zinc finger protein 296
|
|
chr1_-_43389768
Show fit
|
0.13 |
ENST00000372455.4
ENST00000372457.9
ENST00000290663.10
|
MED8
|
mediator complex subunit 8
|
|
chr6_-_119149124
Show fit
|
0.12 |
ENST00000368475.8
|
FAM184A
|
family with sequence similarity 184 member A
|
|
chrX_-_141242512
Show fit
|
0.12 |
ENST00000358993.3
|
SPANXC
|
SPANX family member C
|
|
chrX_-_141698739
Show fit
|
0.12 |
ENST00000370515.3
|
SPANXD
|
SPANX family member D
|
|
chr5_+_162067858
Show fit
|
0.12 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr18_+_34593392
Show fit
|
0.12 |
ENST00000684377.1
|
DTNA
|
dystrobrevin alpha
|
|
chr12_-_56752311
Show fit
|
0.12 |
ENST00000338193.11
ENST00000550770.1
|
PRIM1
|
DNA primase subunit 1
|
|
chr3_-_139678011
Show fit
|
0.11 |
ENST00000646611.1
ENST00000645290.1
ENST00000647257.1
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr2_-_99255107
Show fit
|
0.11 |
ENST00000333017.6
ENST00000626374.2
ENST00000409679.5
ENST00000423306.1
|
LYG2
|
lysozyme g2
|
|
chr1_+_6555301
Show fit
|
0.11 |
ENST00000333172.11
ENST00000351136.7
|
TAS1R1
|
taste 1 receptor member 1
|
|
chrX_+_143025918
Show fit
|
0.11 |
ENST00000446864.1
ENST00000370504.3
|
SPANXN4
|
SPANX family member N4
|
|
chr10_-_29634964
Show fit
|
0.11 |
ENST00000375398.6
ENST00000355867.8
|
SVIL
|
supervillin
|
|
chr17_-_10549652
Show fit
|
0.11 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2
|
|
chr12_+_10505602
Show fit
|
0.10 |
ENST00000322446.3
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B
|
|
chr2_-_230960954
Show fit
|
0.10 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55
|
|
chr2_-_2324968
Show fit
|
0.09 |
ENST00000649641.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr16_+_22008083
Show fit
|
0.09 |
ENST00000542527.7
ENST00000569656.5
ENST00000562695.1
|
MOSMO
|
modulator of smoothened
|
|
chr2_+_227325225
Show fit
|
0.09 |
ENST00000353339.7
|
MFF
|
mitochondrial fission factor
|
|
chr1_+_103750406
Show fit
|
0.08 |
ENST00000370079.3
|
AMY1C
|
amylase alpha 1C
|
|
chr7_+_142715314
Show fit
|
0.08 |
ENST00000390399.3
|
TRBV27
|
T cell receptor beta variable 27
|
|
chr9_-_38705545
Show fit
|
0.08 |
ENST00000637691.1
|
FAM240B
|
family with sequence similarity 240 member B
|
|
chr7_-_150632333
Show fit
|
0.08 |
ENST00000493969.2
ENST00000328902.9
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr11_-_33753394
Show fit
|
0.07 |
ENST00000532057.5
ENST00000531080.5
|
FBXO3
|
F-box protein 3
|
|
chr15_+_21637909
Show fit
|
0.07 |
ENST00000332663.3
|
OR4M2B
|
olfactory receptor family 4 subfamily M member 2B
|
|
chr2_+_21137468
Show fit
|
0.07 |
ENST00000622654.1
|
TDRD15
|
tudor domain containing 15
|
|
chr1_+_172659095
Show fit
|
0.07 |
ENST00000367721.3
ENST00000340030.4
|
FASLG
|
Fas ligand
|
|
chr12_-_68159732
Show fit
|
0.07 |
ENST00000229135.4
|
IFNG
|
interferon gamma
|
|
chr12_-_118359105
Show fit
|
0.07 |
ENST00000541186.5
ENST00000539872.5
|
TAOK3
|
TAO kinase 3
|
|
chr2_+_151409878
Show fit
|
0.06 |
ENST00000453091.6
ENST00000428287.6
ENST00000444746.7
ENST00000243326.9
ENST00000414861.6
|
RIF1
|
replication timing regulatory factor 1
|
|
chr1_+_150067279
Show fit
|
0.06 |
ENST00000643970.1
ENST00000535106.5
ENST00000369128.9
|
VPS45
|
vacuolar protein sorting 45 homolog
|