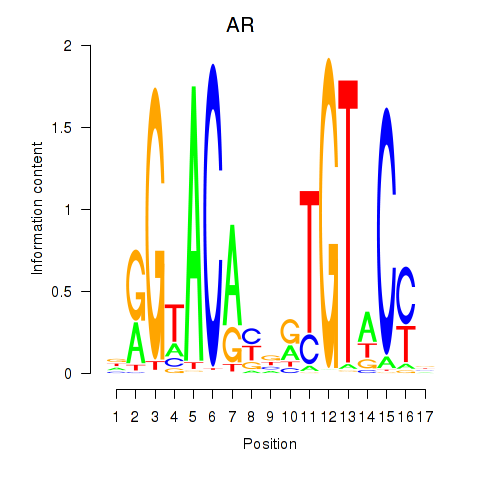
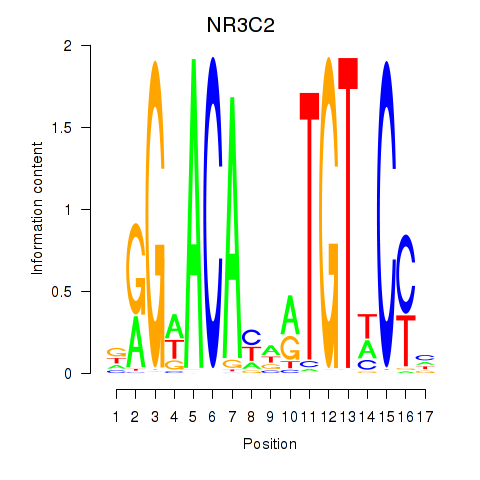
|
chr11_+_18266254
Show fit
|
12.93 |
ENST00000532858.5
ENST00000649195.1
ENST00000356524.9
ENST00000405158.2
|
SAA1
|
serum amyloid A1
|
|
chr12_+_53050179
Show fit
|
11.82 |
ENST00000546602.5
ENST00000552570.5
ENST00000549700.5
|
TNS2
|
tensin 2
|
|
chr12_+_53050014
Show fit
|
9.54 |
ENST00000314250.11
|
TNS2
|
tensin 2
|
|
chr1_+_22653228
Show fit
|
8.02 |
ENST00000509305.6
|
C1QB
|
complement C1q B chain
|
|
chr16_+_56682461
Show fit
|
7.94 |
ENST00000562939.1
ENST00000394485.5
ENST00000567563.1
|
MT1X
ENSG00000259827.1
|
metallothionein 1X
novel transcript
|
|
chr1_+_22653189
Show fit
|
6.60 |
ENST00000432749.6
|
C1QB
|
complement C1q B chain
|
|
chr16_+_56632651
Show fit
|
6.48 |
ENST00000379818.4
ENST00000570233.1
|
MT1M
|
metallothionein 1M
|
|
chr19_+_35139724
Show fit
|
6.43 |
ENST00000588715.5
ENST00000588607.5
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr19_+_35140022
Show fit
|
5.77 |
ENST00000588081.5
ENST00000589121.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr3_-_58577367
Show fit
|
5.30 |
ENST00000464064.5
ENST00000360997.7
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr3_-_58577648
Show fit
|
4.91 |
ENST00000394481.5
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr8_+_24440930
Show fit
|
4.57 |
ENST00000441335.6
ENST00000175238.10
ENST00000380789.5
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr17_-_19387170
Show fit
|
4.49 |
ENST00000395592.6
ENST00000299610.5
|
MFAP4
|
microfibril associated protein 4
|
|
chr19_+_35139440
Show fit
|
4.47 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr2_+_237487239
Show fit
|
4.42 |
ENST00000338530.8
ENST00000264605.8
ENST00000409373.5
|
MLPH
|
melanophilin
|
|
chr18_-_50819982
Show fit
|
4.39 |
ENST00000398439.8
ENST00000431965.6
ENST00000436348.6
|
MRO
|
maestro
|
|
chr14_-_23154422
Show fit
|
4.27 |
ENST00000422941.6
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr19_-_42412347
Show fit
|
4.18 |
ENST00000601189.1
ENST00000599211.1
|
LIPE
|
lipase E, hormone sensitive type
|
|
chrX_+_8464830
Show fit
|
4.16 |
ENST00000453306.4
ENST00000381032.6
ENST00000444481.3
|
VCX3B
|
variable charge X-linked 3B
|
|
chr2_+_102337148
Show fit
|
4.09 |
ENST00000311734.6
ENST00000409584.5
|
IL1RL1
|
interleukin 1 receptor like 1
|
|
chrX_-_6535118
Show fit
|
4.06 |
ENST00000381089.7
ENST00000612369.4
ENST00000398729.1
|
VCX3A
|
variable charge X-linked 3A
|
|
chr16_+_56625775
Show fit
|
4.01 |
ENST00000330439.7
ENST00000568293.1
|
MT1E
|
metallothionein 1E
|
|
chrY_+_14056226
Show fit
|
3.94 |
ENST00000250823.5
|
VCY1B
|
variable charge Y-linked 1B
|
|
chr4_+_74445126
Show fit
|
3.85 |
ENST00000395748.8
|
AREG
|
amphiregulin
|
|
chrY_-_13986473
Show fit
|
3.75 |
ENST00000250825.5
|
VCY
|
variable charge Y-linked
|
|
chr16_+_56608577
Show fit
|
3.58 |
ENST00000245185.6
ENST00000561491.1
|
MT2A
|
metallothionein 2A
|
|
chr17_-_19386785
Show fit
|
3.56 |
ENST00000497081.6
|
MFAP4
|
microfibril associated protein 4
|
|
chrX_+_8465426
Show fit
|
3.52 |
ENST00000381029.4
|
VCX3B
|
variable charge X-linked 3B
|
|
chr4_+_74445302
Show fit
|
3.46 |
ENST00000502307.1
|
AREG
|
amphiregulin
|
|
chr14_-_23154369
Show fit
|
3.43 |
ENST00000453702.5
|
SLC7A8
|
solute carrier family 7 member 8
|
|
chr17_-_8152380
Show fit
|
3.35 |
ENST00000317276.9
|
PER1
|
period circadian regulator 1
|
|
chrX_+_7843088
Show fit
|
2.94 |
ENST00000341408.5
|
VCX
|
variable charge X-linked
|
|
chr16_+_56638659
Show fit
|
2.93 |
ENST00000290705.12
|
MT1A
|
metallothionein 1A
|
|
chr20_-_37158904
Show fit
|
2.80 |
ENST00000417458.5
|
MROH8
|
maestro heat like repeat family member 8
|
|
chr9_-_35563867
Show fit
|
2.68 |
ENST00000399742.7
ENST00000619051.4
|
FAM166B
|
family with sequence similarity 166 member B
|
|
chr15_+_42402375
Show fit
|
2.65 |
ENST00000397200.8
ENST00000569827.5
|
CAPN3
|
calpain 3
|
|
chrX_-_8171267
Show fit
|
2.53 |
ENST00000317103.5
|
VCX2
|
variable charge X-linked 2
|
|
chr13_+_113122791
Show fit
|
2.50 |
ENST00000375559.8
ENST00000409306.5
ENST00000375551.7
|
F10
|
coagulation factor X
|
|
chr7_-_75611702
Show fit
|
2.13 |
ENST00000616821.4
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1
|
|
chr6_-_111605859
Show fit
|
2.09 |
ENST00000651359.1
ENST00000650859.1
ENST00000359831.8
ENST00000368761.11
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr18_+_7754959
Show fit
|
1.98 |
ENST00000400053.8
|
PTPRM
|
protein tyrosine phosphatase receptor type M
|
|
chr4_-_68951763
Show fit
|
1.95 |
ENST00000251566.9
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3
|
|
chr3_-_119677346
Show fit
|
1.81 |
ENST00000484810.5
ENST00000497116.1
ENST00000261070.7
|
COX17
|
cytochrome c oxidase copper chaperone COX17
|
|
chr18_+_49560684
Show fit
|
1.78 |
ENST00000577628.5
|
LIPG
|
lipase G, endothelial type
|
|
chr3_-_119146014
Show fit
|
1.72 |
ENST00000441144.6
ENST00000425327.6
|
IGSF11
|
immunoglobulin superfamily member 11
|
|
chrX_-_81121663
Show fit
|
1.67 |
ENST00000430960.5
ENST00000447319.5
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr8_-_21812320
Show fit
|
1.66 |
ENST00000517328.5
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr10_-_27983103
Show fit
|
1.65 |
ENST00000672841.1
|
ODAD2
|
outer dynein arm docking complex subunit 2
|
|
chr4_+_87006988
Show fit
|
1.56 |
ENST00000307808.10
|
AFF1
|
AF4/FMR2 family member 1
|
|
chr19_-_14835162
Show fit
|
1.46 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5
|
|
chr9_+_128920966
Show fit
|
1.43 |
ENST00000428610.5
ENST00000372592.8
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chrX_-_81121619
Show fit
|
1.41 |
ENST00000373250.7
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr19_-_8343255
Show fit
|
1.39 |
ENST00000330915.7
ENST00000593649.5
ENST00000595639.1
|
KANK3
|
KN motif and ankyrin repeat domains 3
|
|
chr20_-_45547752
Show fit
|
1.38 |
ENST00000409554.1
|
EPPIN
|
epididymal peptidase inhibitor
|
|
chr3_+_119146159
Show fit
|
1.37 |
ENST00000295622.6
|
TEX55
|
testis expressed 55
|
|
chrX_+_80014746
Show fit
|
1.37 |
ENST00000373296.8
|
TBX22
|
T-box transcription factor 22
|
|
chr20_-_45547373
Show fit
|
1.35 |
ENST00000354280.9
|
EPPIN
|
epididymal peptidase inhibitor
|
|
chr22_-_22559073
Show fit
|
1.35 |
ENST00000420709.5
ENST00000398741.5
|
PRAME
|
PRAME nuclear receptor transcriptional regulator
|
|
chr3_+_119146184
Show fit
|
1.27 |
ENST00000460150.1
|
TEX55
|
testis expressed 55
|
|
chr17_+_48841563
Show fit
|
1.26 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2
|
|
chr8_+_61287950
Show fit
|
1.26 |
ENST00000519846.5
ENST00000325897.5
ENST00000523868.2
ENST00000518592.5
|
CLVS1
|
clavesin 1
|
|
chr4_+_87006736
Show fit
|
1.25 |
ENST00000544085.6
|
AFF1
|
AF4/FMR2 family member 1
|
|
chr22_+_22704265
Show fit
|
1.22 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22
|
|
chr20_-_45547420
Show fit
|
1.21 |
ENST00000504988.1
|
EPPIN-WFDC6
|
EPPIN-WFDC6 readthrough
|
|
chr20_-_45547648
Show fit
|
1.20 |
ENST00000651288.1
|
EPPIN-WFDC6
|
EPPIN-WFDC6 readthrough
|
|
chr14_-_21025490
Show fit
|
1.17 |
ENST00000553442.5
ENST00000555869.5
ENST00000556457.5
ENST00000397844.6
ENST00000554415.5
ENST00000556974.5
ENST00000554419.5
ENST00000298687.9
ENST00000397858.5
ENST00000360463.7
ENST00000350792.7
ENST00000397847.6
|
NDRG2
|
NDRG family member 2
|
|
chr16_+_67173971
Show fit
|
1.16 |
ENST00000563258.1
ENST00000568146.1
|
NOL3
|
nucleolar protein 3
|
|
chr8_+_109540075
Show fit
|
1.10 |
ENST00000614147.1
ENST00000337573.10
|
EBAG9
|
estrogen receptor binding site associated antigen 9
|
|
chr6_+_127577168
Show fit
|
1.09 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr4_+_185426234
Show fit
|
1.07 |
ENST00000511138.5
ENST00000511581.5
ENST00000378850.5
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr1_-_159862648
Show fit
|
1.07 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8
|
|
chr7_-_94655993
Show fit
|
1.07 |
ENST00000647110.1
ENST00000647048.1
ENST00000643020.1
ENST00000644682.1
ENST00000646119.1
ENST00000646265.1
ENST00000645445.1
ENST00000647334.1
ENST00000645262.1
ENST00000428696.7
ENST00000647096.1
ENST00000642394.1
ENST00000645725.1
ENST00000647351.1
ENST00000646943.1
ENST00000648936.2
ENST00000642707.1
ENST00000645109.1
ENST00000646489.1
ENST00000642933.1
ENST00000643128.1
ENST00000445866.7
ENST00000645101.1
ENST00000644116.1
ENST00000642441.1
ENST00000646879.1
ENST00000647018.1
|
SGCE
|
sarcoglycan epsilon
|
|
chr15_-_28099293
Show fit
|
1.05 |
ENST00000431101.1
ENST00000445578.5
ENST00000353809.9
ENST00000354638.8
|
OCA2
|
OCA2 melanosomal transmembrane protein
|
|
chr1_+_103561757
Show fit
|
1.03 |
ENST00000435302.5
|
AMY2B
|
amylase alpha 2B
|
|
chr17_+_20579724
Show fit
|
1.03 |
ENST00000661883.1
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15 like 2
|
|
chr5_-_173616588
Show fit
|
1.02 |
ENST00000285908.5
ENST00000311086.9
ENST00000480951.1
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr1_-_10796636
Show fit
|
0.96 |
ENST00000377022.8
ENST00000344008.5
|
CASZ1
|
castor zinc finger 1
|
|
chr16_+_67173935
Show fit
|
0.95 |
ENST00000566871.5
|
NOL3
|
nucleolar protein 3
|
|
chr12_+_5043873
Show fit
|
0.95 |
ENST00000252321.5
|
KCNA5
|
potassium voltage-gated channel subfamily A member 5
|
|
chr11_+_90085950
Show fit
|
0.95 |
ENST00000530464.2
|
UBTFL1
|
upstream binding transcription factor like 1
|
|
chr17_+_39667964
Show fit
|
0.92 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase
|
|
chr10_-_329480
Show fit
|
0.84 |
ENST00000434695.2
|
DIP2C
|
disco interacting protein 2 homolog C
|
|
chr12_+_50925007
Show fit
|
0.84 |
ENST00000332160.5
|
METTL7A
|
methyltransferase like 7A
|
|
chr8_-_144063951
Show fit
|
0.83 |
ENST00000567871.2
|
OPLAH
|
5-oxoprolinase, ATP-hydrolysing
|
|
chr7_-_97024821
Show fit
|
0.83 |
ENST00000648378.1
ENST00000486603.2
|
DLX5
|
distal-less homeobox 5
|
|
chr7_+_20615653
Show fit
|
0.79 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5
|
|
chr4_-_185425941
Show fit
|
0.79 |
ENST00000264689.11
ENST00000505357.1
|
UFSP2
|
UFM1 specific peptidase 2
|
|
chr11_-_114400417
Show fit
|
0.79 |
ENST00000325636.8
ENST00000623205.2
|
C11orf71
|
chromosome 11 open reading frame 71
|
|
chr17_+_81097564
Show fit
|
0.74 |
ENST00000576756.5
|
BAIAP2
|
BAR/IMD domain containing adaptor protein 2
|
|
chr1_-_204149881
Show fit
|
0.73 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2
|
|
chr12_-_10098977
Show fit
|
0.72 |
ENST00000315330.8
ENST00000457018.6
|
CLEC1A
|
C-type lectin domain family 1 member A
|
|
chr1_+_248488589
Show fit
|
0.69 |
ENST00000641363.1
|
OR2T5
|
olfactory receptor family 2 subfamily T member 5
|
|
chr6_+_18387326
Show fit
|
0.69 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B
|
|
chr19_+_15010720
Show fit
|
0.69 |
ENST00000292574.4
|
CCDC105
|
coiled-coil domain containing 105
|
|
chr8_-_22156789
Show fit
|
0.68 |
ENST00000306317.7
|
LGI3
|
leucine rich repeat LGI family member 3
|
|
chr8_-_61689768
Show fit
|
0.64 |
ENST00000517847.6
ENST00000389204.8
ENST00000517661.5
ENST00000517903.5
ENST00000522603.5
ENST00000541428.5
ENST00000522349.5
ENST00000522835.5
ENST00000518306.5
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr18_-_50819764
Show fit
|
0.63 |
ENST00000592966.5
|
MRO
|
maestro
|
|
chr1_-_24143112
Show fit
|
0.63 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
|
chr1_+_75796867
Show fit
|
0.62 |
ENST00000263187.4
|
MSH4
|
mutS homolog 4
|
|
chr8_+_90001448
Show fit
|
0.61 |
ENST00000519410.5
ENST00000522161.5
ENST00000220764.7
ENST00000517761.5
ENST00000520227.1
|
DECR1
|
2,4-dienoyl-CoA reductase 1
|
|
chr13_+_113759219
Show fit
|
0.61 |
ENST00000375353.5
ENST00000488362.5
|
TMEM255B
|
transmembrane protein 255B
|
|
chr12_+_10010627
Show fit
|
0.58 |
ENST00000338896.11
ENST00000396502.5
|
CLEC12B
|
C-type lectin domain family 12 member B
|
|
chr17_-_63773534
Show fit
|
0.58 |
ENST00000403162.7
ENST00000582252.1
ENST00000225726.10
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr14_+_24147474
Show fit
|
0.57 |
ENST00000324103.11
ENST00000559260.5
|
RNF31
|
ring finger protein 31
|
|
chr16_-_67944113
Show fit
|
0.53 |
ENST00000264005.10
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr8_-_48921419
Show fit
|
0.52 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2
|
|
chr16_-_71442033
Show fit
|
0.51 |
ENST00000561754.4
|
TLE7
|
TLE family member 7
|
|
chr12_-_106086809
Show fit
|
0.47 |
ENST00000548902.1
|
NUAK1
|
NUAK family kinase 1
|
|
chr19_+_35250563
Show fit
|
0.47 |
ENST00000599658.1
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr18_+_58862904
Show fit
|
0.47 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532
|
|
chr14_+_21782993
Show fit
|
0.46 |
ENST00000390429.3
|
TRAV7
|
T cell receptor alpha variable 7
|
|
chr17_+_81057615
Show fit
|
0.41 |
ENST00000572073.5
ENST00000573677.5
|
BAIAP2
|
BAR/IMD domain containing adaptor protein 2
|
|
chr4_+_87007175
Show fit
|
0.41 |
ENST00000511722.5
|
AFF1
|
AF4/FMR2 family member 1
|
|
chr7_-_94656160
Show fit
|
0.35 |
ENST00000644609.1
ENST00000643272.1
ENST00000646137.1
ENST00000646098.1
ENST00000643193.1
ENST00000437425.7
ENST00000644551.1
ENST00000415788.3
|
SGCE
|
sarcoglycan epsilon
|
|
chr2_-_201769759
Show fit
|
0.31 |
ENST00000680163.1
ENST00000680188.1
ENST00000679550.1
|
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2
|
|
chrX_-_117985298
Show fit
|
0.30 |
ENST00000469946.5
|
KLHL13
|
kelch like family member 13
|
|
chr12_+_125065427
Show fit
|
0.29 |
ENST00000316519.11
|
AACS
|
acetoacetyl-CoA synthetase
|
|
chr11_-_75669028
Show fit
|
0.29 |
ENST00000304771.8
|
MAP6
|
microtubule associated protein 6
|
|
chr16_+_8642375
Show fit
|
0.28 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22
|
|
chr8_+_7768977
Show fit
|
0.28 |
ENST00000533716.2
|
FAM90A10P
|
family with sequence similarity 90 member A10, pseudogene
|
|
chr8_-_7582653
Show fit
|
0.26 |
ENST00000648435.1
|
FAM90A23P
|
family with sequence similarity 90 member A23, pseudogene
|
|
chr22_+_21567705
Show fit
|
0.26 |
ENST00000342192.9
|
UBE2L3
|
ubiquitin conjugating enzyme E2 L3
|
|
chr11_-_236326
Show fit
|
0.26 |
ENST00000525237.1
ENST00000382743.9
ENST00000532956.5
ENST00000525319.5
ENST00000524564.5
|
SIRT3
|
sirtuin 3
|
|
chr8_+_7715443
Show fit
|
0.25 |
ENST00000648315.1
|
FAM90A14P
|
family with sequence similarity 90 member A14, pseudogene
|
|
chr18_+_49561013
Show fit
|
0.25 |
ENST00000583083.1
|
LIPG
|
lipase G, endothelial type
|
|
chr7_-_94656197
Show fit
|
0.25 |
ENST00000643903.1
ENST00000644122.1
ENST00000447873.6
ENST00000644816.1
ENST00000644375.1
|
SGCE
|
sarcoglycan epsilon
|
|
chr4_+_37891060
Show fit
|
0.23 |
ENST00000261439.9
ENST00000508802.5
ENST00000402522.1
|
TBC1D1
|
TBC1 domain family member 1
|
|
chr3_-_134374891
Show fit
|
0.23 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2
|
|
chr8_+_7730739
Show fit
|
0.20 |
ENST00000648174.1
|
FAM90A16P
|
family with sequence similarity 90 member A16, pseudogene
|
|
chr2_-_75561297
Show fit
|
0.20 |
ENST00000410071.5
ENST00000432649.5
|
EVA1A
|
eva-1 homolog A, regulator of programmed cell death
|
|
chr3_+_13480215
Show fit
|
0.20 |
ENST00000446613.6
ENST00000402259.5
ENST00000402271.5
ENST00000295757.8
ENST00000404548.5
ENST00000404040.5
|
HDAC11
|
histone deacetylase 11
|
|
chr12_-_56454653
Show fit
|
0.15 |
ENST00000652304.1
|
MIP
|
major intrinsic protein of lens fiber
|
|
chr8_-_109603439
Show fit
|
0.15 |
ENST00000533394.5
|
SYBU
|
syntabulin
|
|
chr2_-_241637045
Show fit
|
0.12 |
ENST00000407315.6
|
THAP4
|
THAP domain containing 4
|
|
chr12_+_56221977
Show fit
|
0.11 |
ENST00000447747.5
ENST00000399713.6
|
NABP2
|
nucleic acid binding protein 2
|
|
chr11_+_8911280
Show fit
|
0.10 |
ENST00000530281.5
ENST00000396648.6
ENST00000534147.5
ENST00000529942.1
|
AKIP1
|
A-kinase interacting protein 1
|
|
chr11_+_114400592
Show fit
|
0.10 |
ENST00000541475.5
|
RBM7
|
RNA binding motif protein 7
|
|
chr2_+_190180930
Show fit
|
0.09 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr17_+_43171187
Show fit
|
0.08 |
ENST00000542611.5
ENST00000590996.6
ENST00000589872.1
|
NBR1
|
NBR1 autophagy cargo receptor
|
|
chr4_-_8440712
Show fit
|
0.08 |
ENST00000356406.10
ENST00000413009.6
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr18_+_63542365
Show fit
|
0.05 |
ENST00000269491.6
ENST00000382768.2
|
SERPINB12
|
serpin family B member 12
|
|
chr19_+_55676621
Show fit
|
0.04 |
ENST00000411543.6
|
EPN1
|
epsin 1
|
|
chr8_-_7559712
Show fit
|
0.04 |
ENST00000650288.1
|
FAM90A7P
|
family with sequence similarity 90 member A7, pseudogene
|
|
chr19_+_8053000
Show fit
|
0.03 |
ENST00000390669.7
|
CCL25
|
C-C motif chemokine ligand 25
|
|
chr3_-_24165548
Show fit
|
0.02 |
ENST00000280696.9
|
THRB
|
thyroid hormone receptor beta
|
|
chr22_+_19951503
Show fit
|
0.02 |
ENST00000406520.7
|
COMT
|
catechol-O-methyltransferase
|
|
chr8_+_7746034
Show fit
|
0.01 |
ENST00000650426.1
|
FAM90A17P
|
family with sequence similarity 90 member A17, pseudogene
|
|
chr8_+_7761330
Show fit
|
0.01 |
ENST00000648344.1
|
FAM90A9P
|
family with sequence similarity 90 member A9, pseudogene
|
|
chr8_+_7738387
Show fit
|
0.01 |
ENST00000647775.1
|
FAM90A8P
|
family with sequence similarity 90 member A8, pseudogene
|
|
chr8_+_7723091
Show fit
|
0.00 |
ENST00000647769.1
|
FAM90A18P
|
family with sequence similarity 90 member A18, pseudogene
|
|
chr8_+_7753682
Show fit
|
0.00 |
ENST00000647903.1
|
FAM90A19P
|
family with sequence similarity 90 member A19, pseudogene
|