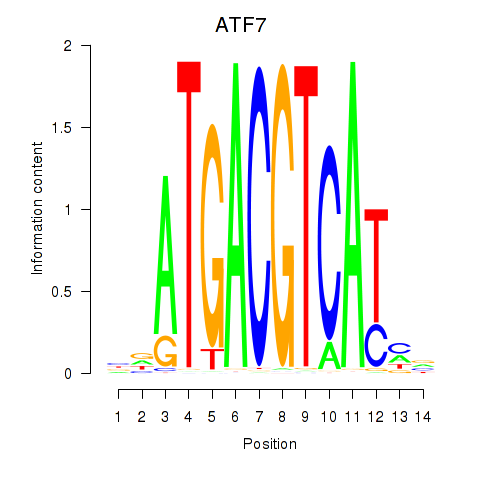
|
chr11_-_77474087
Show fit
|
3.33 |
ENST00000356341.8
|
PAK1
|
p21 (RAC1) activated kinase 1
|
|
chr11_-_77474041
Show fit
|
3.18 |
ENST00000278568.8
|
PAK1
|
p21 (RAC1) activated kinase 1
|
|
chr14_+_92923143
Show fit
|
3.09 |
ENST00000216492.10
ENST00000334654.4
|
CHGA
|
chromogranin A
|
|
chr22_+_31122923
Show fit
|
2.73 |
ENST00000620191.4
ENST00000412277.6
ENST00000412985.5
ENST00000331075.10
ENST00000420017.5
ENST00000400294.6
ENST00000405300.5
ENST00000404390.7
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr11_-_13495984
Show fit
|
2.72 |
ENST00000282091.6
|
PTH
|
parathyroid hormone
|
|
chr11_-_13496018
Show fit
|
2.53 |
ENST00000529816.1
|
PTH
|
parathyroid hormone
|
|
chr22_-_43862480
Show fit
|
2.32 |
ENST00000330884.9
|
SULT4A1
|
sulfotransferase family 4A member 1
|
|
chr17_+_70075215
Show fit
|
2.27 |
ENST00000283936.5
ENST00000615244.4
ENST00000392671.6
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16
|
|
chr19_-_50333504
Show fit
|
2.08 |
ENST00000474951.1
|
KCNC3
|
potassium voltage-gated channel subfamily C member 3
|
|
chr17_+_70075317
Show fit
|
1.79 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16
|
|
chr6_+_150368892
Show fit
|
1.79 |
ENST00000229447.9
ENST00000392256.6
|
IYD
|
iodotyrosine deiodinase
|
|
chr6_+_150368997
Show fit
|
1.78 |
ENST00000392255.7
ENST00000500320.7
ENST00000344419.8
|
IYD
|
iodotyrosine deiodinase
|
|
chr20_+_5911501
Show fit
|
1.69 |
ENST00000378961.9
ENST00000455042.1
|
CHGB
|
chromogranin B
|
|
chr14_+_67619911
Show fit
|
1.52 |
ENST00000261783.4
|
ARG2
|
arginase 2
|
|
chr6_-_26285526
Show fit
|
1.45 |
ENST00000377727.2
|
H4C8
|
H4 clustered histone 8
|
|
chrX_+_111096136
Show fit
|
1.23 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3
|
|
chr4_+_158210194
Show fit
|
1.21 |
ENST00000508243.5
|
TMEM144
|
transmembrane protein 144
|
|
chrX_+_153072454
Show fit
|
1.18 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A
|
|
chrX_+_111096211
Show fit
|
1.16 |
ENST00000372010.5
ENST00000519681.5
|
PAK3
|
p21 (RAC1) activated kinase 3
|
|
chr16_-_68023201
Show fit
|
1.13 |
ENST00000332395.7
|
DDX28
|
DEAD-box helicase 28
|
|
chr14_-_93184840
Show fit
|
1.12 |
ENST00000298894.5
ENST00000556883.1
|
MOAP1
|
modulator of apoptosis 1
|
|
chr15_+_74995520
Show fit
|
1.11 |
ENST00000562327.5
ENST00000568018.5
ENST00000425597.8
ENST00000562212.5
ENST00000567920.5
ENST00000566872.5
ENST00000361900.10
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr11_-_123654581
Show fit
|
1.08 |
ENST00000392770.6
ENST00000530277.5
ENST00000299333.8
|
SCN3B
|
sodium voltage-gated channel beta subunit 3
|
|
chr6_+_56955097
Show fit
|
1.04 |
ENST00000370746.8
ENST00000370748.7
|
BEND6
|
BEN domain containing 6
|
|
chr11_+_66291887
Show fit
|
1.02 |
ENST00000327259.5
|
TMEM151A
|
transmembrane protein 151A
|
|
chr12_-_122896066
Show fit
|
0.99 |
ENST00000267202.7
ENST00000535765.5
|
VPS37B
|
VPS37B subunit of ESCRT-I
|
|
chr4_+_84583037
Show fit
|
0.98 |
ENST00000295887.6
|
CDS1
|
CDP-diacylglycerol synthase 1
|
|
chr4_+_158210444
Show fit
|
0.97 |
ENST00000512481.5
|
TMEM144
|
transmembrane protein 144
|
|
chr1_+_2073462
Show fit
|
0.96 |
ENST00000400921.6
|
PRKCZ
|
protein kinase C zeta
|
|
chr16_-_4538761
Show fit
|
0.95 |
ENST00000567695.6
ENST00000562334.5
ENST00000562579.5
ENST00000563507.5
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr19_+_18612848
Show fit
|
0.93 |
ENST00000262817.8
|
TMEM59L
|
transmembrane protein 59 like
|
|
chr3_+_62318983
Show fit
|
0.92 |
ENST00000232519.9
ENST00000462069.6
ENST00000465142.5
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr17_+_35587239
Show fit
|
0.91 |
ENST00000621914.4
ENST00000621668.4
ENST00000616681.4
ENST00000612035.4
ENST00000610402.5
ENST00000614600.4
ENST00000590432.5
ENST00000612116.5
|
AP2B1
|
adaptor related protein complex 2 subunit beta 1
|
|
chr16_-_4538819
Show fit
|
0.89 |
ENST00000564828.5
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr7_+_30134956
Show fit
|
0.88 |
ENST00000324453.13
ENST00000409688.1
|
MTURN
|
maturin, neural progenitor differentiation regulator homolog
|
|
chr2_+_148875214
Show fit
|
0.87 |
ENST00000435030.6
ENST00000677891.1
ENST00000677843.1
ENST00000678056.1
ENST00000677280.1
|
KIF5C
|
kinesin family member 5C
|
|
chr6_+_26204552
Show fit
|
0.85 |
ENST00000615164.2
|
H4C5
|
H4 clustered histone 5
|
|
chr19_-_2702682
Show fit
|
0.84 |
ENST00000382159.8
|
GNG7
|
G protein subunit gamma 7
|
|
chr2_-_219309484
Show fit
|
0.84 |
ENST00000409251.7
ENST00000451506.5
ENST00000446182.5
|
PTPRN
|
protein tyrosine phosphatase receptor type N
|
|
chr19_-_47113756
Show fit
|
0.84 |
ENST00000253048.10
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chrX_+_153056458
Show fit
|
0.83 |
ENST00000593810.3
|
PNMA3
|
PNMA family member 3
|
|
chr19_-_46428869
Show fit
|
0.83 |
ENST00000617053.3
|
PNMA8C
|
PNMA family member 8C
|
|
chr2_-_240820205
Show fit
|
0.83 |
ENST00000647731.1
ENST00000648680.1
ENST00000649306.1
ENST00000648129.1
ENST00000498729.9
ENST00000320389.12
ENST00000648364.1
ENST00000647885.1
ENST00000404283.9
ENST00000649096.1
|
KIF1A
|
kinesin family member 1A
|
|
chr2_+_37344594
Show fit
|
0.82 |
ENST00000404976.5
ENST00000338415.8
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr7_+_55951793
Show fit
|
0.82 |
ENST00000446692.5
|
NIPSNAP2
|
nipsnap homolog 2
|
|
chr17_+_43025203
Show fit
|
0.81 |
ENST00000587250.4
|
RND2
|
Rho family GTPase 2
|
|
chr1_+_2073986
Show fit
|
0.77 |
ENST00000461106.6
|
PRKCZ
|
protein kinase C zeta
|
|
chr12_-_51173067
Show fit
|
0.77 |
ENST00000549867.5
ENST00000257915.10
|
TFCP2
|
transcription factor CP2
|
|
chr11_-_123654939
Show fit
|
0.76 |
ENST00000657191.1
|
SCN3B
|
sodium voltage-gated channel beta subunit 3
|
|
chr17_-_21043397
Show fit
|
0.74 |
ENST00000584538.1
|
USP22
|
ubiquitin specific peptidase 22
|
|
chr1_-_153945225
Show fit
|
0.74 |
ENST00000368646.6
|
DENND4B
|
DENN domain containing 4B
|
|
chrX_+_153056408
Show fit
|
0.73 |
ENST00000619635.1
|
PNMA3
|
PNMA family member 3
|
|
chr14_+_103629177
Show fit
|
0.73 |
ENST00000348520.10
ENST00000380038.7
ENST00000389744.8
ENST00000557575.5
ENST00000553286.5
ENST00000347839.10
ENST00000555836.5
ENST00000334553.10
ENST00000246489.11
ENST00000634686.1
ENST00000557450.5
ENST00000452929.6
ENST00000554280.5
ENST00000445352.8
|
KLC1
|
kinesin light chain 1
|
|
chr13_-_113974062
Show fit
|
0.72 |
ENST00000636427.3
|
C13orf46
|
chromosome 13 open reading frame 46
|
|
chrX_+_10158448
Show fit
|
0.71 |
ENST00000380829.5
ENST00000421085.7
ENST00000674669.1
ENST00000454850.1
|
CLCN4
|
chloride voltage-gated channel 4
|
|
chr18_+_74496301
Show fit
|
0.71 |
ENST00000579847.5
ENST00000583203.5
ENST00000581513.5
ENST00000324262.9
ENST00000577600.5
ENST00000579583.5
ENST00000584613.5
|
CNDP2
|
carnosine dipeptidase 2
|
|
chr9_-_122264798
Show fit
|
0.70 |
ENST00000417201.4
|
RBM18
|
RNA binding motif protein 18
|
|
chr10_+_122163590
Show fit
|
0.70 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr16_+_29900474
Show fit
|
0.68 |
ENST00000308748.10
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr7_-_73738792
Show fit
|
0.68 |
ENST00000222800.8
ENST00000458339.6
|
ABHD11
|
abhydrolase domain containing 11
|
|
chr18_+_74496234
Show fit
|
0.67 |
ENST00000580672.5
|
CNDP2
|
carnosine dipeptidase 2
|
|
chr17_+_57256727
Show fit
|
0.67 |
ENST00000675656.1
|
MSI2
|
musashi RNA binding protein 2
|
|
chr7_-_31340678
Show fit
|
0.67 |
ENST00000297142.4
|
NEUROD6
|
neuronal differentiation 6
|
|
chr18_+_79069385
Show fit
|
0.66 |
ENST00000426216.6
ENST00000307671.12
ENST00000586672.5
ENST00000586722.5
|
ATP9B
|
ATPase phospholipid transporting 9B (putative)
|
|
chr2_-_219309350
Show fit
|
0.66 |
ENST00000295718.7
|
PTPRN
|
protein tyrosine phosphatase receptor type N
|
|
chr10_-_37976589
Show fit
|
0.66 |
ENST00000302609.8
|
ZNF25
|
zinc finger protein 25
|
|
chr6_-_42746054
Show fit
|
0.65 |
ENST00000372876.2
|
TBCC
|
tubulin folding cofactor C
|
|
chr4_+_112637120
Show fit
|
0.65 |
ENST00000509061.5
ENST00000344442.10
ENST00000508577.5
ENST00000513553.5
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator
|
|
chr1_+_156061142
Show fit
|
0.64 |
ENST00000361084.10
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr11_-_33161502
Show fit
|
0.64 |
ENST00000438862.6
|
CSTF3
|
cleavage stimulation factor subunit 3
|
|
chr6_+_56955353
Show fit
|
0.64 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6
|
|
chr10_+_122163426
Show fit
|
0.64 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr11_+_109422174
Show fit
|
0.63 |
ENST00000327419.7
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr19_+_55485176
Show fit
|
0.63 |
ENST00000205194.5
ENST00000591590.1
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (putative)
|
|
chr17_+_35587478
Show fit
|
0.62 |
ENST00000618940.4
|
AP2B1
|
adaptor related protein complex 2 subunit beta 1
|
|
chr1_+_109984756
Show fit
|
0.62 |
ENST00000393614.8
ENST00000369799.10
|
AHCYL1
|
adenosylhomocysteinase like 1
|
|
chr10_+_97737115
Show fit
|
0.62 |
ENST00000337540.11
ENST00000370613.7
ENST00000370610.7
ENST00000357540.8
ENST00000393677.8
ENST00000423811.3
ENST00000684270.1
ENST00000359980.5
|
ZFYVE27
|
zinc finger FYVE-type containing 27
|
|
chr6_+_125956696
Show fit
|
0.61 |
ENST00000229633.7
|
HINT3
|
histidine triad nucleotide binding protein 3
|
|
chr1_+_27773189
Show fit
|
0.61 |
ENST00000373943.9
ENST00000440806.2
|
STX12
|
syntaxin 12
|
|
chr3_+_62319037
Show fit
|
0.60 |
ENST00000494481.5
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr7_+_140697144
Show fit
|
0.60 |
ENST00000476470.5
ENST00000471136.5
|
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2
|
|
chr3_+_99817818
Show fit
|
0.58 |
ENST00000463526.1
|
CMSS1
|
cms1 ribosomal small subunit homolog
|
|
chr10_+_122163672
Show fit
|
0.58 |
ENST00000369004.7
ENST00000260733.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr12_+_57604812
Show fit
|
0.58 |
ENST00000337737.8
ENST00000548198.5
ENST00000551632.1
|
DTX3
|
deltex E3 ubiquitin ligase 3
|
|
chr2_-_223602284
Show fit
|
0.58 |
ENST00000421386.1
ENST00000305409.3
ENST00000433889.1
|
SCG2
|
secretogranin II
|
|
chr11_-_31817937
Show fit
|
0.57 |
ENST00000638965.1
ENST00000241001.13
ENST00000638914.3
ENST00000379132.8
ENST00000379129.7
|
PAX6
|
paired box 6
|
|
chr1_-_114581589
Show fit
|
0.57 |
ENST00000369541.4
|
BCAS2
|
BCAS2 pre-mRNA processing factor
|
|
chr4_+_141220881
Show fit
|
0.57 |
ENST00000262990.9
ENST00000512809.5
ENST00000503649.5
ENST00000512738.5
|
ZNF330
|
zinc finger protein 330
|
|
chr12_-_51172779
Show fit
|
0.57 |
ENST00000548108.1
ENST00000548115.5
|
TFCP2
|
transcription factor CP2
|
|
chr3_-_48088800
Show fit
|
0.56 |
ENST00000423088.5
|
MAP4
|
microtubule associated protein 4
|
|
chr12_+_57604904
Show fit
|
0.56 |
ENST00000548478.1
|
DTX3
|
deltex E3 ubiquitin ligase 3
|
|
chr18_+_11851404
Show fit
|
0.55 |
ENST00000526991.3
|
CHMP1B
|
charged multivesicular body protein 1B
|
|
chr13_-_31162341
Show fit
|
0.55 |
ENST00000445273.6
ENST00000630972.2
|
HSPH1
|
heat shock protein family H (Hsp110) member 1
|
|
chr4_-_39366342
Show fit
|
0.55 |
ENST00000503784.1
ENST00000349703.7
ENST00000381897.5
|
RFC1
|
replication factor C subunit 1
|
|
chrX_-_7927375
Show fit
|
0.54 |
ENST00000381042.9
|
PNPLA4
|
patatin like phospholipase domain containing 4
|
|
chr3_+_140941792
Show fit
|
0.54 |
ENST00000446041.6
ENST00000324194.12
ENST00000507429.5
|
SLC25A36
|
solute carrier family 25 member 36
|
|
chr5_-_16508990
Show fit
|
0.53 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1
|
|
chr3_+_99817849
Show fit
|
0.53 |
ENST00000421999.8
|
CMSS1
|
cms1 ribosomal small subunit homolog
|
|
chr3_-_9952337
Show fit
|
0.52 |
ENST00000411976.2
ENST00000412055.6
|
PRRT3
|
proline rich transmembrane protein 3
|
|
chr10_+_119819320
Show fit
|
0.52 |
ENST00000631555.1
ENST00000490818.1
|
INPP5F
|
inositol polyphosphate-5-phosphatase F
|
|
chr5_-_16508788
Show fit
|
0.52 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr20_+_45791978
Show fit
|
0.51 |
ENST00000449078.5
ENST00000456939.5
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1
|
|
chr20_+_45791930
Show fit
|
0.51 |
ENST00000372622.8
|
DNTTIP1
|
deoxynucleotidyltransferase terminal interacting protein 1
|
|
chr19_+_35995176
Show fit
|
0.51 |
ENST00000378887.4
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1
|
|
chr5_-_16508858
Show fit
|
0.51 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr8_+_93917045
Show fit
|
0.50 |
ENST00000518573.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chrX_-_135973684
Show fit
|
0.49 |
ENST00000681201.1
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr9_-_137459296
Show fit
|
0.49 |
ENST00000371472.6
ENST00000371475.9
ENST00000371473.7
ENST00000371474.7
ENST00000437259.5
ENST00000265663.12
|
NSMF
|
NMDA receptor synaptonuclear signaling and neuronal migration factor
|
|
chr12_+_57604598
Show fit
|
0.49 |
ENST00000548804.5
ENST00000550596.5
ENST00000551835.5
ENST00000549583.5
|
DTX3
|
deltex E3 ubiquitin ligase 3
|
|
chr17_-_51046868
Show fit
|
0.49 |
ENST00000510283.5
ENST00000510855.1
|
SPAG9
|
sperm associated antigen 9
|
|
chr20_-_17968438
Show fit
|
0.48 |
ENST00000431277.5
|
SNX5
|
sorting nexin 5
|
|
chr12_+_10212483
Show fit
|
0.48 |
ENST00000545859.5
|
GABARAPL1
|
GABA type A receptor associated protein like 1
|
|
chr3_-_108222383
Show fit
|
0.48 |
ENST00000264538.4
|
IFT57
|
intraflagellar transport 57
|
|
chr4_-_169612571
Show fit
|
0.48 |
ENST00000507142.6
ENST00000510533.5
ENST00000439128.6
ENST00000511633.5
ENST00000512193.5
|
NEK1
|
NIMA related kinase 1
|
|
chr16_+_56191728
Show fit
|
0.48 |
ENST00000638705.1
ENST00000262494.12
|
GNAO1
|
G protein subunit alpha o1
|
|
chr16_-_4538469
Show fit
|
0.47 |
ENST00000588381.1
ENST00000563332.6
|
CDIP1
|
cell death inducing p53 target 1
|
|
chr5_-_16508812
Show fit
|
0.47 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr16_+_56191476
Show fit
|
0.47 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1
|
|
chr12_+_112418976
Show fit
|
0.46 |
ENST00000635625.1
|
PTPN11
|
protein tyrosine phosphatase non-receptor type 11
|
|
chr4_+_169660062
Show fit
|
0.45 |
ENST00000507875.5
ENST00000613795.4
|
CLCN3
|
chloride voltage-gated channel 3
|
|
chr16_+_1989949
Show fit
|
0.45 |
ENST00000248121.7
ENST00000618464.1
|
SYNGR3
|
synaptogyrin 3
|
|
chr19_-_50837213
Show fit
|
0.45 |
ENST00000326856.8
|
KLK15
|
kallikrein related peptidase 15
|
|
chr7_+_98106852
Show fit
|
0.45 |
ENST00000297293.6
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr3_+_184338826
Show fit
|
0.45 |
ENST00000453072.5
|
FAM131A
|
family with sequence similarity 131 member A
|
|
chr4_-_112636858
Show fit
|
0.44 |
ENST00000503172.5
ENST00000505019.6
ENST00000309071.9
|
ZGRF1
|
zinc finger GRF-type containing 1
|
|
chr3_-_154324416
Show fit
|
0.44 |
ENST00000329463.9
|
DHX36
|
DEAH-box helicase 36
|
|
chr16_+_81037187
Show fit
|
0.44 |
ENST00000564241.5
ENST00000565237.1
|
ATMIN
|
ATM interactor
|
|
chr20_-_17968500
Show fit
|
0.43 |
ENST00000419004.2
|
SNX5
|
sorting nexin 5
|
|
chr1_+_248095184
Show fit
|
0.43 |
ENST00000358120.3
ENST00000641893.1
|
OR2L13
|
olfactory receptor family 2 subfamily L member 13
|
|
chr7_+_23181994
Show fit
|
0.43 |
ENST00000410002.7
ENST00000258742.10
ENST00000413919.1
|
NUP42
|
nucleoporin 42
|
|
chr5_-_16508951
Show fit
|
0.43 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1
|
|
chr14_+_21846534
Show fit
|
0.43 |
ENST00000390434.3
|
TRAV8-2
|
T cell receptor alpha variable 8-2
|
|
chr8_+_93916849
Show fit
|
0.43 |
ENST00000520614.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr11_-_33161461
Show fit
|
0.43 |
ENST00000323959.9
ENST00000431742.2
ENST00000524827.6
|
CSTF3
|
cleavage stimulation factor subunit 3
|
|
chr3_-_52278620
Show fit
|
0.43 |
ENST00000296490.8
|
WDR82
|
WD repeat domain 82
|
|
chr3_+_140941901
Show fit
|
0.42 |
ENST00000453248.6
|
SLC25A36
|
solute carrier family 25 member 36
|
|
chr3_-_44761596
Show fit
|
0.42 |
ENST00000296121.6
|
KIAA1143
|
KIAA1143
|
|
chr3_-_52278321
Show fit
|
0.42 |
ENST00000469000.5
|
WDR82
|
WD repeat domain 82
|
|
chr19_+_57320461
Show fit
|
0.41 |
ENST00000321545.5
|
ZNF543
|
zinc finger protein 543
|
|
chr9_-_97922487
Show fit
|
0.41 |
ENST00000455506.1
ENST00000375117.8
ENST00000375119.8
ENST00000611338.4
|
TRMO
|
tRNA methyltransferase O
|
|
chr17_+_63774144
Show fit
|
0.41 |
ENST00000389924.7
ENST00000359353.9
|
DDX42
|
DEAD-box helicase 42
|
|
chr6_+_37433197
Show fit
|
0.41 |
ENST00000455891.5
ENST00000373451.9
|
CMTR1
|
cap methyltransferase 1
|
|
chr19_-_55063081
Show fit
|
0.40 |
ENST00000415061.8
|
RDH13
|
retinol dehydrogenase 13
|
|
chr7_+_140696956
Show fit
|
0.40 |
ENST00000460088.5
ENST00000472695.5
|
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2
|
|
chr2_-_189762755
Show fit
|
0.40 |
ENST00000520350.1
ENST00000521630.1
ENST00000264151.10
ENST00000517895.5
|
OSGEPL1
|
O-sialoglycoprotein endopeptidase like 1
|
|
chr16_+_19067606
Show fit
|
0.40 |
ENST00000321998.10
|
COQ7
|
coenzyme Q7, hydroxylase
|
|
chr5_+_139675471
Show fit
|
0.40 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5
|
|
chr2_+_219544002
Show fit
|
0.40 |
ENST00000421791.1
ENST00000373883.4
ENST00000451952.1
|
TMEM198
|
transmembrane protein 198
|
|
chr1_+_2073688
Show fit
|
0.40 |
ENST00000495347.5
|
PRKCZ
|
protein kinase C zeta
|
|
chr16_+_19067989
Show fit
|
0.40 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7, hydroxylase
|
|
chr8_-_22109381
Show fit
|
0.40 |
ENST00000613958.1
ENST00000611621.2
|
NUDT18
|
nudix hydrolase 18
|
|
chr7_+_140696696
Show fit
|
0.39 |
ENST00000247866.9
ENST00000464566.5
|
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2
|
|
chr3_+_44729596
Show fit
|
0.39 |
ENST00000396048.6
ENST00000620116.5
|
ZNF501
|
zinc finger protein 501
|
|
chr17_+_31094880
Show fit
|
0.39 |
ENST00000487476.5
ENST00000356175.7
|
NF1
|
neurofibromin 1
|
|
chr5_-_138875290
Show fit
|
0.39 |
ENST00000521094.2
ENST00000274711.7
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr7_+_152759744
Show fit
|
0.38 |
ENST00000377776.7
ENST00000256001.13
ENST00000397282.2
|
ACTR3B
|
actin related protein 3B
|
|
chr5_+_113513674
Show fit
|
0.38 |
ENST00000161863.9
ENST00000515883.5
|
YTHDC2
|
YTH domain containing 2
|
|
chr19_+_50476496
Show fit
|
0.38 |
ENST00000597426.1
ENST00000376918.7
ENST00000334976.11
ENST00000598585.1
|
EMC10
|
ER membrane protein complex subunit 10
|
|
chr3_-_108222362
Show fit
|
0.38 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57
|
|
chr18_-_12656716
Show fit
|
0.38 |
ENST00000462226.1
ENST00000497844.6
ENST00000309836.9
ENST00000453447.6
|
SPIRE1
|
spire type actin nucleation factor 1
|
|
chr10_-_14838561
Show fit
|
0.37 |
ENST00000378442.5
|
CDNF
|
cerebral dopamine neurotrophic factor
|
|
chr6_-_127459364
Show fit
|
0.37 |
ENST00000487331.2
ENST00000483725.8
|
KIAA0408
|
KIAA0408
|
|
chr12_+_2812638
Show fit
|
0.37 |
ENST00000228799.7
|
ITFG2
|
integrin alpha FG-GAP repeat containing 2
|
|
chr1_-_244863085
Show fit
|
0.37 |
ENST00000440865.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr7_+_90346679
Show fit
|
0.37 |
ENST00000417207.5
ENST00000222511.11
|
GTPBP10
|
GTP binding protein 10
|
|
chr3_-_187670385
Show fit
|
0.37 |
ENST00000287641.4
|
SST
|
somatostatin
|
|
chr17_-_63773534
Show fit
|
0.37 |
ENST00000403162.7
ENST00000582252.1
ENST00000225726.10
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr7_+_140696665
Show fit
|
0.37 |
ENST00000476279.5
ENST00000461457.1
ENST00000465506.5
|
NDUFB2
|
NADH:ubiquinone oxidoreductase subunit B2
|
|
chrX_-_7927701
Show fit
|
0.36 |
ENST00000537427.5
ENST00000444736.5
ENST00000442940.1
|
PNPLA4
|
patatin like phospholipase domain containing 4
|
|
chr11_-_3797490
Show fit
|
0.36 |
ENST00000397004.8
ENST00000397007.8
ENST00000532475.1
ENST00000324932.12
|
NUP98
|
nucleoporin 98 and 96 precursor
|
|
chr7_+_152025632
Show fit
|
0.36 |
ENST00000430044.7
ENST00000431668.1
ENST00000446096.5
ENST00000423337.5
|
GALNT11
|
polypeptide N-acetylgalactosaminyltransferase 11
|
|
chr19_+_48619291
Show fit
|
0.35 |
ENST00000340932.7
ENST00000601712.5
ENST00000600537.5
|
SPHK2
|
sphingosine kinase 2
|
|
chr3_-_154324446
Show fit
|
0.35 |
ENST00000308361.10
ENST00000496811.6
|
DHX36
|
DEAH-box helicase 36
|
|
chr4_+_145098269
Show fit
|
0.35 |
ENST00000502586.5
ENST00000296577.9
|
ABCE1
|
ATP binding cassette subfamily E member 1
|
|
chr16_+_3018390
Show fit
|
0.35 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A
|
|
chr7_+_142554828
Show fit
|
0.35 |
ENST00000611787.1
|
TRBV11-3
|
T cell receptor beta variable 11-3
|
|
chr12_+_10212867
Show fit
|
0.34 |
ENST00000545047.5
ENST00000266458.10
ENST00000629504.1
ENST00000543602.5
ENST00000545887.1
|
GABARAPL1
|
GABA type A receptor associated protein like 1
|
|
chr8_+_97775829
Show fit
|
0.34 |
ENST00000517924.5
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr12_-_6663083
Show fit
|
0.34 |
ENST00000467678.5
ENST00000493873.1
ENST00000412586.6
ENST00000423703.6
ENST00000444704.5
ENST00000341550.9
|
ING4
|
inhibitor of growth family member 4
|
|
chr19_-_48044037
Show fit
|
0.34 |
ENST00000293255.3
|
CABP5
|
calcium binding protein 5
|
|
chrX_-_47619850
Show fit
|
0.34 |
ENST00000295987.13
ENST00000340666.5
|
SYN1
|
synapsin I
|
|
chr14_+_93184951
Show fit
|
0.34 |
ENST00000283534.4
ENST00000557574.1
|
TMEM251
ENSG00000259066.5
|
transmembrane protein 251
novel protein
|
|
chr9_-_128881922
Show fit
|
0.33 |
ENST00000320665.10
ENST00000436267.7
ENST00000302586.8
ENST00000651925.1
|
KYAT1
ENSG00000286112.1
|
kynurenine aminotransferase 1
kynurenine aminotransferase 1
|
|
chr4_-_48134200
Show fit
|
0.33 |
ENST00000264316.9
|
TXK
|
TXK tyrosine kinase
|
|
chr17_+_63773863
Show fit
|
0.33 |
ENST00000578681.5
ENST00000583590.5
|
DDX42
|
DEAD-box helicase 42
|
|
chr7_+_90346665
Show fit
|
0.33 |
ENST00000257659.12
|
GTPBP10
|
GTP binding protein 10
|
|
chr8_+_30156359
Show fit
|
0.33 |
ENST00000520829.5
ENST00000221114.8
|
DCTN6
|
dynactin subunit 6
|
|
chr7_+_142492121
Show fit
|
0.33 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6
|
|
chr17_+_27294076
Show fit
|
0.33 |
ENST00000581440.5
ENST00000583742.1
ENST00000579733.5
ENST00000583193.5
ENST00000581185.5
ENST00000427287.6
ENST00000262394.7
ENST00000348811.6
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr5_-_138875362
Show fit
|
0.33 |
ENST00000518785.1
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr7_+_75493625
Show fit
|
0.33 |
ENST00000625065.3
|
SPDYE5
|
speedy/RINGO cell cycle regulator family member E5
|
|
chr6_-_16761447
Show fit
|
0.33 |
ENST00000244769.8
ENST00000436367.6
|
ATXN1
|
ataxin 1
|
|
chr7_+_142462882
Show fit
|
0.32 |
ENST00000454561.2
|
TRBV5-4
|
T cell receptor beta variable 5-4
|
|
chr17_-_45133213
Show fit
|
0.32 |
ENST00000538093.1
ENST00000590644.5
|
PLCD3
|
phospholipase C delta 3
|
|
chr9_-_34665985
Show fit
|
0.32 |
ENST00000416454.5
ENST00000544078.2
ENST00000421828.7
ENST00000423809.5
|
ENSG00000187186.15
|
novel protein
|
|
chr19_-_38912177
Show fit
|
0.32 |
ENST00000571838.2
|
CCER2
|
coiled-coil glutamate rich protein 2
|
|
chr3_-_195543308
Show fit
|
0.31 |
ENST00000618156.5
|
PPP1R2
|
protein phosphatase 1 regulatory inhibitor subunit 2
|
|
chr3_+_141368497
Show fit
|
0.31 |
ENST00000321464.7
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chrX_+_134807217
Show fit
|
0.31 |
ENST00000482240.5
ENST00000475361.1
ENST00000645433.2
ENST00000494709.1
|
PABIR3
|
PABIR family member 3
|
|
chr17_-_7252482
Show fit
|
0.31 |
ENST00000572043.5
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|