Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
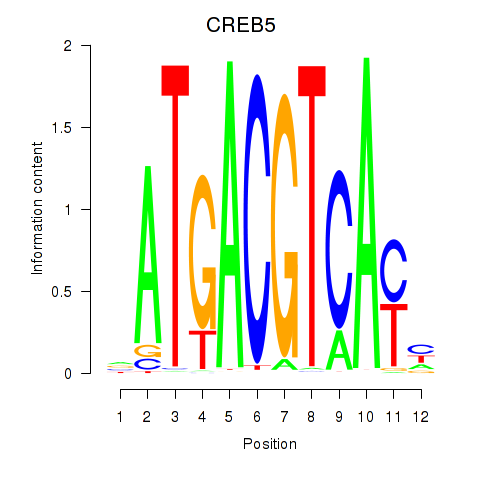
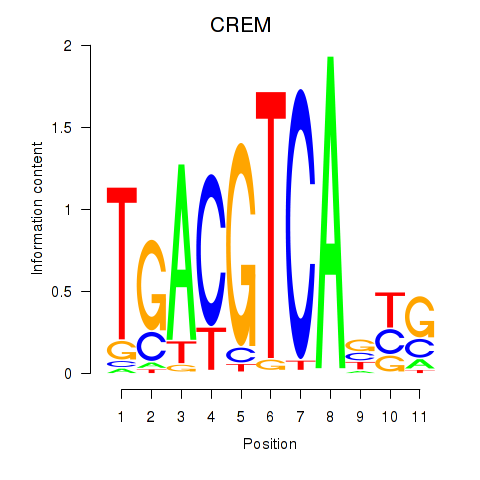
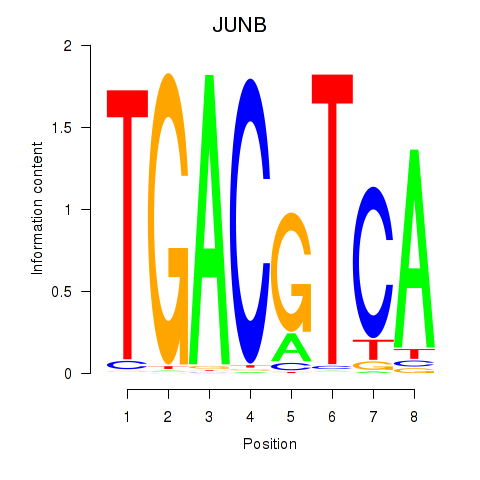
Results for CREB5_CREM_JUNB
Z-value: 3.00
Transcription factors associated with CREB5_CREM_JUNB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB5
|
ENSG00000146592.17 | CREB5 |
|
CREM
|
ENSG00000095794.19 | CREM |
|
JUNB
|
ENSG00000171223.6 | JUNB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB5 | hg38_v1_chr7_+_28409344_28409392 | 0.52 | 2.1e-03 | Click! |
| JUNB | hg38_v1_chr19_+_12791470_12791494 | -0.44 | 1.2e-02 | Click! |
| CREM | hg38_v1_chr10_+_35195124_35195148 | 0.43 | 1.4e-02 | Click! |
Activity profile of CREB5_CREM_JUNB motif
Sorted Z-values of CREB5_CREM_JUNB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB5_CREM_JUNB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.9 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 4.1 | 20.4 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 3.9 | 11.8 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 3.2 | 9.6 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 2.7 | 8.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 2.5 | 7.6 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 2.5 | 7.4 | GO:0036323 | vascular endothelial growth factor receptor-1 signaling pathway(GO:0036323) |
| 2.4 | 7.3 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 2.2 | 11.0 | GO:0030070 | insulin processing(GO:0030070) |
| 2.1 | 27.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.6 | 14.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 1.5 | 4.6 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 1.1 | 4.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.1 | 7.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.1 | 21.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.0 | 4.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 1.0 | 5.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 1.0 | 3.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 1.0 | 3.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 1.0 | 11.9 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.0 | 5.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.9 | 3.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.9 | 2.7 | GO:1901388 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.9 | 2.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.8 | 3.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.8 | 2.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.8 | 3.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) |
| 0.8 | 11.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 6.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.7 | 10.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 2.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.7 | 2.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.7 | 10.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 3.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.6 | 1.9 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 27.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.6 | 4.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.6 | 6.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.6 | 0.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.6 | 2.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.6 | 1.7 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.6 | 6.8 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.6 | 6.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 4.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 7.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 5.8 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.5 | 2.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.5 | 2.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.5 | 2.0 | GO:2000724 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.5 | 7.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.5 | 2.9 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) |
| 0.5 | 1.4 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 1.8 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.4 | 2.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.4 | 1.7 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.4 | 6.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 3.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.4 | 1.7 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 1.7 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 4.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.4 | 4.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 | 3.9 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.4 | 6.5 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.4 | 1.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.4 | 3.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.4 | 4.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.4 | 2.6 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.4 | 1.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 6.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.4 | 3.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 1.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.4 | 4.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 | 2.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 19.0 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 11.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.4 | 3.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 5.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.4 | 4.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 1.7 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.3 | 6.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 4.4 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.3 | 1.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 18.8 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.3 | 9.9 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.3 | 0.9 | GO:1901491 | axial mesoderm formation(GO:0048320) negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 4.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.3 | 5.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.3 | 3.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.3 | 12.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.3 | 7.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 6.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 2.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 2.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.4 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 2.6 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.3 | 5.7 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 | 17.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.3 | 3.6 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.3 | 3.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 0.8 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.3 | 0.8 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.3 | 1.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 1.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 1.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 1.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.3 | 8.1 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 7.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.2 | 5.4 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.2 | 2.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 1.2 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.2 | 2.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.2 | 6.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 10.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 2.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.2 | 6.8 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.2 | 0.7 | GO:1903537 | meiotic spindle elongation(GO:0051232) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 1.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.2 | 10.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 3.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 13.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 2.8 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 3.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.6 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 0.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 | 0.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 1.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.2 | 0.4 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 1.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.5 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.2 | 1.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.0 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.2 | 2.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 3.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.8 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.2 | 0.7 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 1.3 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.2 | 1.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 8.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.2 | 7.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.4 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.2 | 8.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.8 | GO:0060283 | growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 0.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 2.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 1.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.3 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 3.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.9 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 3.9 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.1 | 0.6 | GO:0060734 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 4.4 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 2.5 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 1.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.5 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 1.0 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.3 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 | 5.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 0.9 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 2.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 2.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.2 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 2.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 3.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 2.9 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.8 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 2.9 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 5.0 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 0.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.5 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 4.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 4.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.4 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.4 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 1.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 1.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 3.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 2.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 6.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.9 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.1 | 0.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.6 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 1.8 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 0.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.9 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.5 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 3.3 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.1 | 1.4 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.4 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.1 | 1.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 0.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 4.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 5.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 3.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 7.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 3.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.5 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.1 | 2.9 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 2.5 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.9 | GO:0015820 | leucine transport(GO:0015820) |
| 0.1 | 1.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.7 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 1.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.6 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 0.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.3 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.0 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 9.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 1.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 3.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 1.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.8 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.1 | 0.2 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 3.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) spinal cord association neuron differentiation(GO:0021527) regulation of cell fate specification(GO:0042659) |
| 0.0 | 1.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 4.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.5 | GO:0061436 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 5.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 1.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 7.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.7 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 1.1 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 1.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.8 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 3.1 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 3.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 2.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 1.4 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.6 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:1902963 | modulation of age-related behavioral decline(GO:0090647) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.5 | GO:0010664 | negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 3.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 5.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 3.0 | GO:0007044 | cell-substrate junction assembly(GO:0007044) |
| 0.0 | 0.1 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.3 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.8 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.1 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 1.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 2.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 11.5 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0022028 | tangential migration from the subventricular zone to the olfactory bulb(GO:0022028) |
| 0.0 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.2 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.2 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.0 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.0 | 2.4 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.7 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.8 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 16.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.6 | 17.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.3 | 6.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.1 | 20.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.9 | 2.7 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.8 | 4.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.8 | 10.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.6 | 5.8 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 12.5 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.6 | 2.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.6 | 1.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.6 | 7.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 8.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 2.5 | GO:1990742 | microvesicle(GO:1990742) |
| 0.5 | 3.0 | GO:0044393 | microspike(GO:0044393) |
| 0.5 | 9.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 8.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 1.4 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.4 | 11.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 18.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 1.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 3.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 10.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 9.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 1.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 22.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 4.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 18.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 6.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 2.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 0.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 9.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 7.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 2.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 4.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 3.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.2 | 3.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 2.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 1.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.2 | 2.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 2.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 1.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 5.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 2.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 9.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 19.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 13.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 12.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 6.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 7.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 2.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 9.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 5.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 22.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 5.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 22.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 13.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 4.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 9.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 36.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 6.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 6.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 16.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 2.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 4.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 6.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 2.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 5.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 4.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 1.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 12.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 5.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 6.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 7.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 16.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 3.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 5.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 2.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 3.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.7 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 5.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.8 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 2.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 14.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 6.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.5 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.0 | 10.2 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 5.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 2.5 | 7.4 | GO:0036332 | VEGF-A-activated receptor activity(GO:0036326) VEGF-B-activated receptor activity(GO:0036327) placental growth factor-activated receptor activity(GO:0036332) |
| 1.7 | 8.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 1.5 | 6.2 | GO:0004605 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.5 | 12.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.3 | 11.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.3 | 5.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 1.2 | 21.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.1 | 27.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.0 | 3.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 1.0 | 18.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.9 | 2.7 | GO:0002135 | CTP binding(GO:0002135) |
| 0.8 | 10.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.7 | 5.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.7 | 10.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 6.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 4.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.7 | 2.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 9.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 3.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.6 | 6.3 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.6 | 16.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.6 | 2.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.6 | 7.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 2.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.6 | 14.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.5 | 3.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 3.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.5 | 1.5 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.4 | 1.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.4 | 3.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.4 | 2.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 2.9 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.4 | 2.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 13.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 15.0 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.3 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 3.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 10.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 0.7 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.3 | 1.3 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) L-phenylalanine aminotransferase activity(GO:0070546) |
| 0.3 | 17.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 4.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.3 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 2.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 3.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 8.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 5.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 4.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 9.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 13.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 2.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 3.0 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 1.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.9 | GO:0010465 | neurotrophin receptor activity(GO:0005030) nerve growth factor receptor activity(GO:0010465) neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 0.2 | 2.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 4.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 1.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 4.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 4.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 1.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 1.6 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.3 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 4.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 8.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 6.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 10.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 12.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 3.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 0.9 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.2 | 0.9 | GO:0050610 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.2 | 5.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 7.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 5.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 3.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 7.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 2.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 6.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 12.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 4.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 21.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 3.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 4.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 16.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.8 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 4.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 18.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 2.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 31.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 1.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 2.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 5.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 6.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 8.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 8.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 13.3 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.2 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.1 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 7.2 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 3.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 23.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.2 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 4.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 1.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 2.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 9.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 13.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.0 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 3.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 3.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 7.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 5.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 2.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 3.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.5 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 3.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 7.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 4.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0001132 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 17.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 7.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 11.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 4.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 15.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 2.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 7.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 14.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 15.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 8.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 8.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 5.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 3.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 2.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 11.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 4.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.0 | 27.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.9 | 31.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 23.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 4.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 7.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 14.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 7.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 8.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 7.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.2 | 11.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 8.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 2.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 5.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 5.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 8.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 6.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 4.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 4.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 2.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 6.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 5.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 11.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 6.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 3.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 2.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 2.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 4.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 3.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 9.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 8.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 4.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 14.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.0 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 5.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |