|
chr19_+_4909430
Show fit
|
4.08 |
ENST00000620565.4
ENST00000613817.4
ENST00000624301.3
ENST00000650932.1
|
UHRF1
|
ubiquitin like with PHD and ring finger domains 1
|
|
chr2_+_173354820
Show fit
|
3.83 |
ENST00000347703.7
ENST00000410101.7
ENST00000410019.3
ENST00000306721.8
|
CDCA7
|
cell division cycle associated 7
|
|
chr1_-_25905989
Show fit
|
3.16 |
ENST00000399728.5
|
STMN1
|
stathmin 1
|
|
chr1_-_25906411
Show fit
|
3.06 |
ENST00000455785.7
|
STMN1
|
stathmin 1
|
|
chr20_-_33686371
Show fit
|
3.02 |
ENST00000343380.6
|
E2F1
|
E2F transcription factor 1
|
|
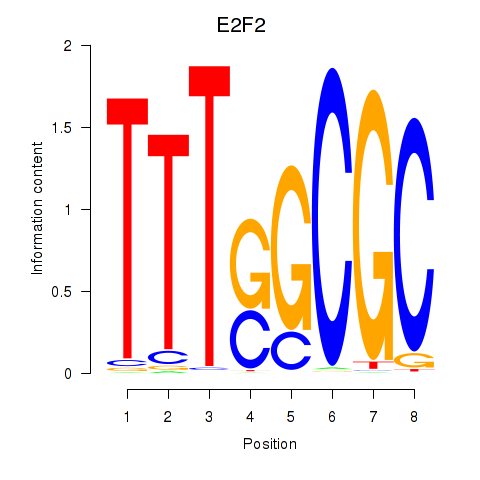
chr1_-_23531206
Show fit
|
2.78 |
ENST00000361729.3
|
E2F2
|
E2F transcription factor 2
|
|
chr1_-_25906931
Show fit
|
2.63 |
ENST00000357865.6
|
STMN1
|
stathmin 1
|
|
chr10_+_110567666
Show fit
|
2.56 |
ENST00000361804.5
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr15_+_32615501
Show fit
|
2.49 |
ENST00000361627.8
ENST00000567348.5
ENST00000563864.5
ENST00000543522.5
|
ARHGAP11A
|
Rho GTPase activating protein 11A
|
|
chr20_-_37095985
Show fit
|
2.47 |
ENST00000344359.7
ENST00000373664.8
|
RBL1
|
RB transcriptional corepressor like 1
|
|
chr3_+_127598400
Show fit
|
2.47 |
ENST00000265056.12
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr22_+_35400115
Show fit
|
2.39 |
ENST00000382011.9
ENST00000216122.9
ENST00000416905.1
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr1_-_25906457
Show fit
|
2.36 |
ENST00000426559.6
|
STMN1
|
stathmin 1
|
|
chr10_-_73874568
Show fit
|
2.01 |
ENST00000322635.7
ENST00000322680.7
ENST00000394762.7
ENST00000680035.1
|
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma
|
|
chr9_-_111038037
Show fit
|
1.95 |
ENST00000374431.7
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr10_-_73874502
Show fit
|
1.93 |
ENST00000372765.5
ENST00000351293.7
ENST00000441192.2
ENST00000423381.6
|
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma
|
|
chr1_-_35769928
Show fit
|
1.85 |
ENST00000373220.7
ENST00000520551.1
|
CLSPN
|
claspin
|
|
chr7_+_94656325
Show fit
|
1.83 |
ENST00000482108.1
ENST00000488574.5
ENST00000612748.1
ENST00000613043.1
|
PEG10
|
paternally expressed 10
|
|
chr9_-_111038425
Show fit
|
1.82 |
ENST00000441240.1
ENST00000683809.1
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr10_-_73874461
Show fit
|
1.78 |
ENST00000305762.11
|
CAMK2G
|
calcium/calmodulin dependent protein kinase II gamma
|
|
chr1_-_120069616
Show fit
|
1.73 |
ENST00000652302.1
ENST00000652737.1
ENST00000256646.7
|
NOTCH2
|
notch receptor 2
|
|
chr20_+_25407657
Show fit
|
1.72 |
ENST00000262460.5
|
GINS1
|
GINS complex subunit 1
|
|
chr8_+_93916882
Show fit
|
1.70 |
ENST00000297598.5
ENST00000520728.5
ENST00000518107.5
ENST00000396200.3
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr12_-_44876294
Show fit
|
1.69 |
ENST00000429094.7
ENST00000551601.5
ENST00000549027.5
ENST00000452445.6
|
NELL2
|
neural EGFL like 2
|
|
chr2_+_15940537
Show fit
|
1.65 |
ENST00000281043.4
ENST00000638417.1
|
MYCN
|
MYCN proto-oncogene, bHLH transcription factor
|
|
chr16_-_2980406
Show fit
|
1.64 |
ENST00000431515.6
ENST00000574385.5
ENST00000576268.1
ENST00000574730.5
ENST00000262300.13
ENST00000575632.5
ENST00000573944.5
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr15_+_40695129
Show fit
|
1.57 |
ENST00000423169.6
ENST00000557850.5
ENST00000382643.7
ENST00000267868.8
ENST00000645673.2
|
RAD51
|
RAD51 recombinase
|
|
chr22_+_41699492
Show fit
|
1.57 |
ENST00000401548.8
ENST00000540833.1
|
MEI1
|
meiotic double-stranded break formation protein 1
|
|
chr3_-_48188356
Show fit
|
1.55 |
ENST00000351231.7
ENST00000437972.1
ENST00000302506.8
|
CDC25A
|
cell division cycle 25A
|
|
chr2_-_17753792
Show fit
|
1.53 |
ENST00000448223.7
ENST00000621152.4
ENST00000351948.8
|
SMC6
|
structural maintenance of chromosomes 6
|
|
chr9_-_111038061
Show fit
|
1.51 |
ENST00000358883.8
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr2_+_27275466
Show fit
|
1.47 |
ENST00000404433.5
ENST00000406962.1
|
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma
|
|
chr8_-_97277890
Show fit
|
1.45 |
ENST00000322128.5
|
TSPYL5
|
TSPY like 5
|
|
chr20_+_2101807
Show fit
|
1.43 |
ENST00000381482.8
|
STK35
|
serine/threonine kinase 35
|
|
chr2_-_234497035
Show fit
|
1.42 |
ENST00000390645.2
ENST00000339728.6
|
ARL4C
|
ADP ribosylation factor like GTPase 4C
|
|
chr1_-_35769958
Show fit
|
1.42 |
ENST00000251195.9
ENST00000318121.8
|
CLSPN
|
claspin
|
|
chr2_-_27275340
Show fit
|
1.39 |
ENST00000424577.5
ENST00000426569.1
|
SLC30A3
|
solute carrier family 30 member 3
|
|
chr5_-_80654956
Show fit
|
1.32 |
ENST00000439211.7
|
DHFR
|
dihydrofolate reductase
|
|
chr22_-_38570167
Show fit
|
1.29 |
ENST00000216024.7
ENST00000616615.4
|
DMC1
|
DNA meiotic recombinase 1
|
|
chr5_-_80654552
Show fit
|
1.29 |
ENST00000511032.5
ENST00000504396.1
ENST00000505337.5
|
DHFR
|
dihydrofolate reductase
|
|
chr10_+_94545852
Show fit
|
1.29 |
ENST00000394036.5
ENST00000394045.5
|
HELLS
|
helicase, lymphoid specific
|
|
chr19_-_56840661
Show fit
|
1.29 |
ENST00000649735.1
ENST00000593695.5
ENST00000599577.5
ENST00000594389.1
ENST00000648694.1
ENST00000326441.15
ENST00000649680.1
ENST00000649876.1
ENST00000650632.1
ENST00000650102.1
ENST00000647621.1
ENST00000598410.5
ENST00000649233.1
ENST00000593711.6
ENST00000629319.2
ENST00000599935.5
|
PEG3
ZIM2
|
paternally expressed 3
zinc finger imprinted 2
|
|
chr11_+_65041203
Show fit
|
1.29 |
ENST00000652489.1
ENST00000674184.1
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr3_+_16884942
Show fit
|
1.27 |
ENST00000615277.5
|
PLCL2
|
phospholipase C like 2
|
|
chr13_-_41194485
Show fit
|
1.26 |
ENST00000379483.4
|
KBTBD7
|
kelch repeat and BTB domain containing 7
|
|
chr10_+_94545777
Show fit
|
1.25 |
ENST00000348459.10
ENST00000419900.5
ENST00000630929.2
|
HELLS
|
helicase, lymphoid specific
|
|
chr2_+_27275429
Show fit
|
1.22 |
ENST00000420191.5
ENST00000296097.8
|
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma
|
|
chr16_-_2980282
Show fit
|
1.22 |
ENST00000572619.1
ENST00000574415.5
ENST00000440027.6
ENST00000572059.1
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr2_-_27263034
Show fit
|
1.22 |
ENST00000233535.9
|
SLC30A3
|
solute carrier family 30 member 3
|
|
chr11_+_65040895
Show fit
|
1.20 |
ENST00000531072.1
ENST00000398846.6
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr20_-_5950463
Show fit
|
1.17 |
ENST00000203001.7
ENST00000453074.6
|
TRMT6
|
tRNA methyltransferase 6
|
|
chr15_-_66356672
Show fit
|
1.16 |
ENST00000261881.9
|
TIPIN
|
TIMELESS interacting protein
|
|
chr3_+_44761765
Show fit
|
1.16 |
ENST00000326047.9
|
KIF15
|
kinesin family member 15
|
|
chr22_+_41998780
Show fit
|
1.15 |
ENST00000328823.13
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr12_-_132687307
Show fit
|
1.14 |
ENST00000535270.5
ENST00000320574.10
|
POLE
|
DNA polymerase epsilon, catalytic subunit
|
|
chrX_+_102651366
Show fit
|
1.13 |
ENST00000415986.5
ENST00000444152.5
ENST00000361600.9
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr15_+_40695423
Show fit
|
1.13 |
ENST00000526763.6
ENST00000532743.6
|
RAD51
|
RAD51 recombinase
|
|
chr2_-_96034916
Show fit
|
1.11 |
ENST00000359548.8
ENST00000439254.1
ENST00000453542.5
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr2_-_17753757
Show fit
|
1.11 |
ENST00000446852.5
|
SMC6
|
structural maintenance of chromosomes 6
|
|
chr6_+_89829931
Show fit
|
1.06 |
ENST00000551025.4
|
CASP8AP2
|
caspase 8 associated protein 2
|
|
chr6_+_89829888
Show fit
|
1.06 |
ENST00000552401.5
ENST00000419040.6
|
CASP8AP2
|
caspase 8 associated protein 2
|
|
chr7_-_100101333
Show fit
|
1.06 |
ENST00000303887.10
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr14_-_49688201
Show fit
|
1.06 |
ENST00000553805.2
ENST00000554396.5
ENST00000216367.10
|
POLE2
|
DNA polymerase epsilon 2, accessory subunit
|
|
chr1_+_120723939
Show fit
|
1.05 |
ENST00000624419.2
ENST00000652763.1
ENST00000620612.5
|
NOTCH2NLR
NBPF26
|
notch 2 N-terminal like R (pseudogene)
NBPF member 26
|
|
chr10_-_73247241
Show fit
|
1.04 |
ENST00000372950.6
|
DNAJC9
|
DnaJ heat shock protein family (Hsp40) member C9
|
|
chr9_-_120877167
Show fit
|
1.03 |
ENST00000373896.8
ENST00000312189.10
|
PHF19
|
PHD finger protein 19
|
|
chr4_+_153344671
Show fit
|
1.02 |
ENST00000240488.8
|
MND1
|
meiotic nuclear divisions 1
|
|
chr11_-_74731385
Show fit
|
1.02 |
ENST00000622063.4
ENST00000376332.8
|
CHRDL2
|
chordin like 2
|
|
chr4_+_70993542
Show fit
|
1.02 |
ENST00000504730.5
ENST00000504952.1
ENST00000286648.10
|
DCK
|
deoxycytidine kinase
|
|
chr3_-_56683218
Show fit
|
1.00 |
ENST00000355628.9
ENST00000683822.1
|
TASOR
|
transcription activation suppressor
|
|
chr12_-_44875647
Show fit
|
0.98 |
ENST00000395487.6
|
NELL2
|
neural EGFL like 2
|
|
chr19_-_36214645
Show fit
|
0.98 |
ENST00000304116.10
|
ZNF565
|
zinc finger protein 565
|
|
chr7_+_120950763
Show fit
|
0.98 |
ENST00000339121.9
ENST00000315870.10
ENST00000445699.5
|
ING3
|
inhibitor of growth family member 3
|
|
chr22_-_38570118
Show fit
|
0.98 |
ENST00000439567.5
|
DMC1
|
DNA meiotic recombinase 1
|
|
chr7_-_158704740
Show fit
|
0.98 |
ENST00000409339.3
ENST00000356309.8
ENST00000409423.5
|
NCAPG2
|
non-SMC condensin II complex subunit G2
|
|
chr3_-_133661896
Show fit
|
0.97 |
ENST00000260810.10
|
TOPBP1
|
DNA topoisomerase II binding protein 1
|
|
chr2_-_218568291
Show fit
|
0.97 |
ENST00000418019.5
ENST00000454775.5
ENST00000338465.5
ENST00000415516.5
ENST00000258399.8
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr7_-_148884159
Show fit
|
0.94 |
ENST00000478654.5
ENST00000460911.5
ENST00000350995.6
|
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit
|
|
chr19_-_10194898
Show fit
|
0.92 |
ENST00000359526.9
ENST00000679103.1
ENST00000676610.1
ENST00000678804.1
ENST00000679313.1
ENST00000677946.1
|
DNMT1
|
DNA methyltransferase 1
|
|
chr9_+_79571767
Show fit
|
0.91 |
ENST00000376544.7
|
TLE4
|
TLE family member 4, transcriptional corepressor
|
|
chr20_-_62407274
Show fit
|
0.91 |
ENST00000279101.9
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr12_+_27780224
Show fit
|
0.89 |
ENST00000381271.7
|
KLHL42
|
kelch like family member 42
|
|
chr5_+_43120883
Show fit
|
0.88 |
ENST00000515326.5
ENST00000682664.1
|
ZNF131
|
zinc finger protein 131
|
|
chr2_-_135876382
Show fit
|
0.87 |
ENST00000264156.3
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr5_-_11904417
Show fit
|
0.85 |
ENST00000304623.13
|
CTNND2
|
catenin delta 2
|
|
chr1_+_151070740
Show fit
|
0.84 |
ENST00000368918.8
|
GABPB2
|
GA binding protein transcription factor subunit beta 2
|
|
chr2_+_27275461
Show fit
|
0.83 |
ENST00000402462.5
|
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma
|
|
chr2_+_46941199
Show fit
|
0.82 |
ENST00000319190.11
ENST00000394850.6
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr10_-_56361235
Show fit
|
0.81 |
ENST00000373944.8
ENST00000361148.6
ENST00000395405.5
|
ZWINT
|
ZW10 interacting kinetochore protein
|
|
chrX_+_24693879
Show fit
|
0.81 |
ENST00000379068.8
ENST00000677890.1
ENST00000379059.7
|
POLA1
|
DNA polymerase alpha 1, catalytic subunit
|
|
chr8_-_119855838
Show fit
|
0.81 |
ENST00000313655.5
|
DSCC1
|
DNA replication and sister chromatid cohesion 1
|
|
chr17_-_7294592
Show fit
|
0.80 |
ENST00000007699.10
|
YBX2
|
Y-box binding protein 2
|
|
chr8_+_47961028
Show fit
|
0.80 |
ENST00000650216.1
|
MCM4
|
minichromosome maintenance complex component 4
|
|
chr17_-_59106801
Show fit
|
0.80 |
ENST00000393065.6
ENST00000262294.12
ENST00000393066.7
|
TRIM37
|
tripartite motif containing 37
|
|
chr4_-_129093454
Show fit
|
0.78 |
ENST00000281142.10
ENST00000511426.5
|
SCLT1
|
sodium channel and clathrin linker 1
|
|
chr3_+_10026409
Show fit
|
0.78 |
ENST00000287647.7
ENST00000676013.1
ENST00000675286.1
ENST00000419585.5
|
FANCD2
|
FA complementation group D2
|
|
chrX_+_102651476
Show fit
|
0.77 |
ENST00000537097.2
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr9_-_35079923
Show fit
|
0.77 |
ENST00000378643.8
ENST00000448890.1
|
FANCG
|
FA complementation group G
|
|
chr2_+_17754116
Show fit
|
0.77 |
ENST00000381254.7
ENST00000524465.5
ENST00000532257.1
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease
|
|
chr11_-_74731148
Show fit
|
0.77 |
ENST00000263671.9
ENST00000528789.1
|
CHRDL2
|
chordin like 2
|
|
chr16_+_88803776
Show fit
|
0.76 |
ENST00000301019.9
|
CDT1
|
chromatin licensing and DNA replication factor 1
|
|
chr8_+_47960883
Show fit
|
0.76 |
ENST00000648407.1
ENST00000649838.1
ENST00000649919.1
ENST00000262105.6
ENST00000649973.1
|
MCM4
|
minichromosome maintenance complex component 4
|
|
chrX_+_76427666
Show fit
|
0.75 |
ENST00000361470.4
|
MAGEE1
|
MAGE family member E1
|
|
chr2_+_10123171
Show fit
|
0.73 |
ENST00000615152.5
|
RRM2
|
ribonucleotide reductase regulatory subunit M2
|
|
chr17_+_30831944
Show fit
|
0.73 |
ENST00000321990.5
|
ATAD5
|
ATPase family AAA domain containing 5
|
|
chr7_-_6009019
Show fit
|
0.73 |
ENST00000382321.5
ENST00000265849.12
|
PMS2
|
PMS1 homolog 2, mismatch repair system component
|
|
chr19_+_49361783
Show fit
|
0.70 |
ENST00000594268.1
|
DKKL1
|
dickkopf like acrosomal protein 1
|
|
chr19_-_10568968
Show fit
|
0.70 |
ENST00000393599.3
|
CDKN2D
|
cyclin dependent kinase inhibitor 2D
|
|
chr19_-_13906062
Show fit
|
0.70 |
ENST00000591586.5
ENST00000346736.6
|
BRME1
|
break repair meiotic recombinase recruitment factor 1
|
|
chr7_-_148884266
Show fit
|
0.70 |
ENST00000483967.5
ENST00000320356.7
|
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit
|
|
chr5_+_43121504
Show fit
|
0.70 |
ENST00000509156.5
ENST00000508259.5
ENST00000306938.8
|
ZNF131
|
zinc finger protein 131
|
|
chr3_+_14947680
Show fit
|
0.69 |
ENST00000435454.5
ENST00000323373.10
|
NR2C2
|
nuclear receptor subfamily 2 group C member 2
|
|
chr1_-_146229000
Show fit
|
0.69 |
ENST00000612520.2
ENST00000579793.6
ENST00000362074.7
|
NBPF10
NOTCH2NLA
|
NBPF member 10
notch 2 N-terminal like A
|
|
chr19_-_10569022
Show fit
|
0.69 |
ENST00000335766.2
|
CDKN2D
|
cyclin dependent kinase inhibitor 2D
|
|
chr1_+_53459391
Show fit
|
0.69 |
ENST00000371445.3
|
DMRTB1
|
DMRT like family B with proline rich C-terminal 1
|
|
chr10_+_100912955
Show fit
|
0.69 |
ENST00000370271.7
ENST00000238961.9
ENST00000370269.3
ENST00000609386.1
|
SLF2
|
SMC5-SMC6 complex localization factor 2
|
|
chr1_-_180502536
Show fit
|
0.68 |
ENST00000367595.4
|
ACBD6
|
acyl-CoA binding domain containing 6
|
|
chr1_-_202710428
Show fit
|
0.68 |
ENST00000367268.5
|
SYT2
|
synaptotagmin 2
|
|
chr15_+_44427793
Show fit
|
0.67 |
ENST00000558966.5
ENST00000559793.5
ENST00000558968.1
|
CTDSPL2
|
CTD small phosphatase like 2
|
|
chr6_+_31739948
Show fit
|
0.67 |
ENST00000375755.8
ENST00000425703.5
ENST00000375750.9
ENST00000375703.7
ENST00000375740.7
|
MSH5
|
mutS homolog 5
|
|
chr17_+_44656429
Show fit
|
0.67 |
ENST00000409122.7
|
MEIOC
|
meiosis specific with coiled-coil domain
|
|
chr2_+_47783172
Show fit
|
0.66 |
ENST00000540021.6
|
MSH6
|
mutS homolog 6
|
|
chr7_-_100428657
Show fit
|
0.66 |
ENST00000360951.8
ENST00000398027.6
ENST00000684423.1
ENST00000472716.1
|
ZCWPW1
|
zinc finger CW-type and PWWP domain containing 1
|
|
chr20_+_5950630
Show fit
|
0.65 |
ENST00000378883.5
ENST00000378896.7
ENST00000378886.6
ENST00000610722.4
|
MCM8
|
minichromosome maintenance 8 homologous recombination repair factor
|
|
chr6_+_24775413
Show fit
|
0.65 |
ENST00000378054.6
ENST00000620958.4
ENST00000476555.5
|
GMNN
|
geminin DNA replication inhibitor
|
|
chr7_-_6009072
Show fit
|
0.65 |
ENST00000642456.1
ENST00000642292.1
ENST00000441476.6
|
PMS2
|
PMS1 homolog 2, mismatch repair system component
|
|
chr7_-_105876575
Show fit
|
0.64 |
ENST00000318724.8
ENST00000419735.8
|
ATXN7L1
|
ataxin 7 like 1
|
|
chr12_-_44875980
Show fit
|
0.64 |
ENST00000548826.5
|
NELL2
|
neural EGFL like 2
|
|
chr5_+_94618653
Show fit
|
0.63 |
ENST00000265140.10
ENST00000504099.5
|
SLF1
|
SMC5-SMC6 complex localization factor 1
|
|
chr12_+_80707625
Show fit
|
0.63 |
ENST00000228641.4
|
MYF6
|
myogenic factor 6
|
|
chr6_-_27132750
Show fit
|
0.62 |
ENST00000607124.1
ENST00000339812.3
|
H2BC11
|
H2B clustered histone 11
|
|
chr9_+_97983332
Show fit
|
0.62 |
ENST00000339399.5
|
ANP32B
|
acidic nuclear phosphoprotein 32 family member B
|
|
chr12_-_48105808
Show fit
|
0.62 |
ENST00000448372.5
|
SENP1
|
SUMO specific peptidase 1
|
|
chrX_-_63755032
Show fit
|
0.62 |
ENST00000624538.2
ENST00000636276.1
ENST00000624843.3
ENST00000671907.1
ENST00000624210.3
ENST00000374870.8
ENST00000635967.1
ENST00000253401.10
ENST00000672194.1
ENST00000637723.2
ENST00000637417.1
ENST00000637520.1
ENST00000374872.4
ENST00000636926.1
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9
|
|
chr19_-_10333512
Show fit
|
0.62 |
ENST00000617231.5
ENST00000611074.4
ENST00000615032.4
|
RAVER1
|
ribonucleoprotein, PTB binding 1
|
|
chr5_+_134526100
Show fit
|
0.62 |
ENST00000395003.5
|
JADE2
|
jade family PHD finger 2
|
|
chrX_+_47232866
Show fit
|
0.61 |
ENST00000218348.7
ENST00000377107.7
|
USP11
|
ubiquitin specific peptidase 11
|
|
chr4_-_129093377
Show fit
|
0.60 |
ENST00000506368.5
ENST00000439369.6
ENST00000503215.5
|
SCLT1
|
sodium channel and clathrin linker 1
|
|
chr1_-_151146611
Show fit
|
0.60 |
ENST00000341697.7
ENST00000368914.8
|
SEMA6C
|
semaphorin 6C
|
|
chr16_-_85688912
Show fit
|
0.59 |
ENST00000253462.8
|
GINS2
|
GINS complex subunit 2
|
|
chr5_+_43121596
Show fit
|
0.59 |
ENST00000505606.6
ENST00000509634.5
ENST00000509341.5
|
ZNF131
|
zinc finger protein 131
|
|
chr2_+_218568558
Show fit
|
0.58 |
ENST00000627282.2
ENST00000542068.5
|
CNOT9
|
CCR4-NOT transcription complex subunit 9
|
|
chr12_-_12696171
Show fit
|
0.58 |
ENST00000651487.1
ENST00000332427.6
ENST00000540796.5
|
GPR19
|
G protein-coupled receptor 19
|
|
chr7_+_120951116
Show fit
|
0.58 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family member 3
|
|
chrX_-_136767322
Show fit
|
0.57 |
ENST00000370620.5
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6
|
|
chr22_-_28741783
Show fit
|
0.56 |
ENST00000439200.5
ENST00000405598.5
ENST00000398017.3
ENST00000649563.1
ENST00000650281.1
ENST00000425190.7
ENST00000404276.6
ENST00000650233.1
ENST00000348295.7
ENST00000382580.6
|
CHEK2
|
checkpoint kinase 2
|
|
chr3_-_14178615
Show fit
|
0.56 |
ENST00000511155.1
|
XPC
|
XPC complex subunit, DNA damage recognition and repair factor
|
|
chr6_+_27133032
Show fit
|
0.55 |
ENST00000359193.3
|
H2AC11
|
H2A clustered histone 11
|
|
chr15_-_70096604
Show fit
|
0.55 |
ENST00000559048.5
ENST00000560939.5
ENST00000440567.7
ENST00000557907.5
ENST00000558379.5
ENST00000559929.5
|
TLE3
|
TLE family member 3, transcriptional corepressor
|
|
chr2_+_47403116
Show fit
|
0.55 |
ENST00000645506.1
ENST00000406134.5
ENST00000233146.7
|
MSH2
|
mutS homolog 2
|
|
chr2_+_218568865
Show fit
|
0.55 |
ENST00000295701.9
|
CNOT9
|
CCR4-NOT transcription complex subunit 9
|
|
chr2_-_144517506
Show fit
|
0.54 |
ENST00000431672.4
ENST00000558170.6
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr12_-_48106042
Show fit
|
0.53 |
ENST00000551798.1
ENST00000549518.6
|
SENP1
|
SUMO specific peptidase 1
|
|
chr3_+_134795248
Show fit
|
0.53 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1
|
|
chr16_+_1989949
Show fit
|
0.53 |
ENST00000248121.7
ENST00000618464.1
|
SYNGR3
|
synaptogyrin 3
|
|
chr17_-_42745025
Show fit
|
0.53 |
ENST00000592492.5
ENST00000585893.5
ENST00000593214.5
ENST00000590078.5
ENST00000428826.7
ENST00000586382.5
ENST00000415827.6
ENST00000592743.5
ENST00000586089.5
|
EZH1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit
|
|
chr13_+_45120493
Show fit
|
0.53 |
ENST00000340473.8
|
GTF2F2
|
general transcription factor IIF subunit 2
|
|
chr1_+_35573308
Show fit
|
0.52 |
ENST00000373235.4
|
TFAP2E
|
transcription factor AP-2 epsilon
|
|
chr1_-_27914513
Show fit
|
0.52 |
ENST00000313433.11
ENST00000373912.8
ENST00000444045.1
|
RPA2
|
replication protein A2
|
|
chr1_+_149390612
Show fit
|
0.52 |
ENST00000650865.1
ENST00000652191.1
ENST00000621744.4
|
NOTCH2NLC
ENSG00000286185.1
|
notch 2 N-terminal like C
novel protein, identical to neuroblastoma breakpoint family, member 19 NBPF19
|
|
chr16_+_2969548
Show fit
|
0.51 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member 4
|
|
chr1_-_47314089
Show fit
|
0.51 |
ENST00000360380.7
ENST00000371877.8
ENST00000447475.7
|
STIL
|
STIL centriolar assembly protein
|
|
chr9_+_104094260
Show fit
|
0.51 |
ENST00000286398.11
ENST00000440179.5
ENST00000374793.8
|
SMC2
|
structural maintenance of chromosomes 2
|
|
chrX_+_132023294
Show fit
|
0.51 |
ENST00000481105.5
ENST00000354719.10
ENST00000394334.7
ENST00000394335.6
|
STK26
|
serine/threonine kinase 26
|
|
chr8_-_94553444
Show fit
|
0.51 |
ENST00000297591.10
ENST00000421249.2
|
VIRMA
|
vir like m6A methyltransferase associated
|
|
chr1_-_91021996
Show fit
|
0.51 |
ENST00000337393.10
|
ZNF644
|
zinc finger protein 644
|
|
chr15_-_40108861
Show fit
|
0.50 |
ENST00000354670.9
ENST00000559701.5
ENST00000557870.1
ENST00000558774.5
|
BMF
|
Bcl2 modifying factor
|
|
chr2_+_237798764
Show fit
|
0.50 |
ENST00000316997.9
|
RBM44
|
RNA binding motif protein 44
|
|
chrX_+_123962040
Show fit
|
0.50 |
ENST00000435215.5
|
STAG2
|
stromal antigen 2
|
|
chr2_+_47783082
Show fit
|
0.50 |
ENST00000614496.4
ENST00000622629.4
ENST00000234420.11
ENST00000616033.4
ENST00000673637.1
|
MSH6
|
mutS homolog 6
|
|
chr3_+_44761831
Show fit
|
0.49 |
ENST00000481166.6
|
KIF15
|
kinesin family member 15
|
|
chr6_+_24774925
Show fit
|
0.49 |
ENST00000356509.7
ENST00000230056.8
|
GMNN
|
geminin DNA replication inhibitor
|
|
chr4_-_129093548
Show fit
|
0.49 |
ENST00000503401.1
|
SCLT1
|
sodium channel and clathrin linker 1
|
|
chr2_-_144517663
Show fit
|
0.49 |
ENST00000427902.5
ENST00000462355.2
ENST00000470879.5
ENST00000409487.7
ENST00000435831.5
ENST00000630572.2
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_-_53046031
Show fit
|
0.49 |
ENST00000482396.5
ENST00000394752.8
|
SFMBT1
|
Scm like with four mbt domains 1
|
|
chr9_+_104094557
Show fit
|
0.48 |
ENST00000374787.7
|
SMC2
|
structural maintenance of chromosomes 2
|
|
chr13_-_41132728
Show fit
|
0.47 |
ENST00000379485.2
|
KBTBD6
|
kelch repeat and BTB domain containing 6
|
|
chr10_+_60778331
Show fit
|
0.47 |
ENST00000519078.6
ENST00000316629.8
ENST00000395284.8
|
CDK1
|
cyclin dependent kinase 1
|
|
chr5_+_36876731
Show fit
|
0.47 |
ENST00000282516.13
ENST00000448238.2
|
NIPBL
|
NIPBL cohesin loading factor
|
|
chr19_+_2236816
Show fit
|
0.46 |
ENST00000221494.10
|
SF3A2
|
splicing factor 3a subunit 2
|
|
chr6_+_83859640
Show fit
|
0.46 |
ENST00000369679.4
ENST00000369681.10
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr17_+_29390326
Show fit
|
0.46 |
ENST00000261716.8
|
TAOK1
|
TAO kinase 1
|
|
chr4_+_153344633
Show fit
|
0.46 |
ENST00000622785.4
|
MND1
|
meiotic nuclear divisions 1
|
|
chr1_-_151146643
Show fit
|
0.46 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C
|
|
chr12_-_48852133
Show fit
|
0.46 |
ENST00000552512.5
ENST00000551468.1
|
DDX23
|
DEAD-box helicase 23
|
|
chr19_-_12919256
Show fit
|
0.45 |
ENST00000293695.8
|
SYCE2
|
synaptonemal complex central element protein 2
|
|
chr8_-_131040890
Show fit
|
0.45 |
ENST00000286355.10
|
ADCY8
|
adenylate cyclase 8
|
|
chr15_+_44427591
Show fit
|
0.45 |
ENST00000558791.5
ENST00000260327.9
|
CTDSPL2
|
CTD small phosphatase like 2
|
|
chr11_-_64972023
Show fit
|
0.45 |
ENST00000530444.5
|
MAJIN
|
membrane anchored junction protein
|
|
chr16_+_30395400
Show fit
|
0.44 |
ENST00000320159.2
ENST00000613509.2
|
ZNF48
|
zinc finger protein 48
|
|
chr10_+_59176600
Show fit
|
0.44 |
ENST00000373880.9
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein like
|
|
chr13_+_97976632
Show fit
|
0.43 |
ENST00000490680.5
ENST00000403772.8
|
IPO5
|
importin 5
|
|
chr1_+_65992389
Show fit
|
0.43 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B
|
|
chr8_+_93916849
Show fit
|
0.43 |
ENST00000520614.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr1_+_25272502
Show fit
|
0.43 |
ENST00000328664.9
|
RHD
|
Rh blood group D antigen
|
|
chr17_-_44503369
Show fit
|
0.43 |
ENST00000585614.1
ENST00000591680.6
|
GPATCH8
|
G-patch domain containing 8
|
|
chr3_+_14947568
Show fit
|
0.42 |
ENST00000413118.5
ENST00000425241.5
|
NR2C2
|
nuclear receptor subfamily 2 group C member 2
|
|
chr1_-_42335189
Show fit
|
0.42 |
ENST00000361776.5
ENST00000445886.5
ENST00000361346.6
|
FOXJ3
|
forkhead box J3
|
|
chr13_-_78603539
Show fit
|
0.42 |
ENST00000377208.7
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr7_-_100101915
Show fit
|
0.42 |
ENST00000621318.4
ENST00000343023.10
|
MCM7
|
minichromosome maintenance complex component 7
|