Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for GCM2
Z-value: 0.87
Transcription factors associated with GCM2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
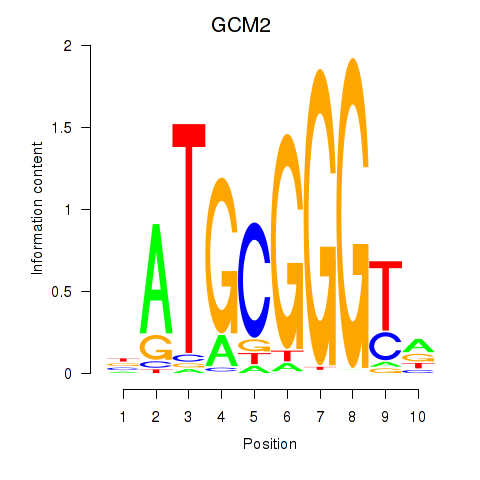
GCM2
|
ENSG00000124827.7 | GCM2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GCM2 | hg38_v1_chr6_-_10882037_10882048 | 0.21 | 2.4e-01 | Click! |
Activity profile of GCM2 motif
Sorted Z-values of GCM2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GCM2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_44325421 | 2.17 |
ENST00000395747.6
ENST00000347193.8 ENST00000346990.8 ENST00000258682.10 ENST00000353625.8 ENST00000421607.1 ENST00000424197.5 |
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chr3_+_50674896 | 2.12 |
ENST00000266037.10
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr2_-_192194908 | 1.64 |
ENST00000392314.5
ENST00000272771.10 |
TMEFF2
|
transmembrane protein with EGF like and two follistatin like domains 2 |
| chr7_+_71132123 | 1.55 |
ENST00000333538.10
|
GALNT17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr9_+_100578071 | 1.49 |
ENST00000307584.6
|
CAVIN4
|
caveolae associated protein 4 |
| chr16_+_71358713 | 1.48 |
ENST00000349553.9
ENST00000302628.9 ENST00000562305.5 |
CALB2
|
calbindin 2 |
| chr11_-_118152775 | 1.21 |
ENST00000324727.9
|
SCN4B
|
sodium voltage-gated channel beta subunit 4 |
| chr11_-_12008584 | 1.13 |
ENST00000534511.5
ENST00000683431.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr19_+_35292145 | 1.13 |
ENST00000595791.5
ENST00000597035.5 ENST00000537831.2 ENST00000392213.8 |
MAG
|
myelin associated glycoprotein |
| chr7_-_44325490 | 1.05 |
ENST00000350811.7
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chr5_+_138439020 | 1.04 |
ENST00000378339.7
ENST00000254901.9 ENST00000506158.5 |
REEP2
|
receptor accessory protein 2 |
| chr9_-_127873462 | 1.03 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr14_+_104579757 | 1.02 |
ENST00000331952.6
ENST00000557649.6 |
C14orf180
|
chromosome 14 open reading frame 180 |
| chr1_+_99646025 | 1.01 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr19_+_35292125 | 1.01 |
ENST00000600291.5
ENST00000361922.8 |
MAG
|
myelin associated glycoprotein |
| chr3_+_35639515 | 0.99 |
ENST00000684406.1
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr12_+_4909895 | 0.98 |
ENST00000638821.1
ENST00000382545.5 |
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1 potassium voltage-gated channel subfamily A member 1 |
| chr11_-_12009199 | 0.97 |
ENST00000533813.5
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr13_-_36131286 | 0.96 |
ENST00000255448.8
ENST00000379892.4 |
DCLK1
|
doublecortin like kinase 1 |
| chr7_+_141074038 | 0.94 |
ENST00000565468.6
ENST00000610315.1 |
TMEM178B
|
transmembrane protein 178B |
| chr13_-_36131352 | 0.90 |
ENST00000360631.8
|
DCLK1
|
doublecortin like kinase 1 |
| chr3_+_156291020 | 0.89 |
ENST00000302490.12
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr7_-_44325653 | 0.87 |
ENST00000440254.6
|
CAMK2B
|
calcium/calmodulin dependent protein kinase II beta |
| chr14_+_75069632 | 0.86 |
ENST00000439583.2
ENST00000554763.2 ENST00000524913.3 ENST00000525046.2 ENST00000674086.1 ENST00000526130.2 ENST00000674094.1 ENST00000532198.2 |
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr15_+_43517590 | 0.86 |
ENST00000300231.6
|
MAP1A
|
microtubule associated protein 1A |
| chr11_-_12009082 | 0.86 |
ENST00000396505.7
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr19_-_19515542 | 0.85 |
ENST00000585580.4
|
TSSK6
|
testis specific serine kinase 6 |
| chr14_-_77271200 | 0.85 |
ENST00000298352.5
|
NGB
|
neuroglobin |
| chr8_+_144504131 | 0.84 |
ENST00000394955.3
|
GPT
|
glutamic--pyruvic transaminase |
| chr3_-_142963663 | 0.82 |
ENST00000340634.6
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr1_-_182672882 | 0.81 |
ENST00000367557.8
ENST00000258302.8 |
RGS8
|
regulator of G protein signaling 8 |
| chr17_-_3893040 | 0.80 |
ENST00000158166.5
ENST00000348335.7 |
CAMKK1
|
calcium/calmodulin dependent protein kinase kinase 1 |
| chr3_+_35639589 | 0.78 |
ENST00000428373.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr2_+_219434825 | 0.77 |
ENST00000312358.12
|
SPEG
|
striated muscle enriched protein kinase |
| chr19_+_18612848 | 0.77 |
ENST00000262817.8
|
TMEM59L
|
transmembrane protein 59 like |
| chr15_-_93089192 | 0.76 |
ENST00000329082.11
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr11_-_12009134 | 0.73 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr1_-_99005003 | 0.73 |
ENST00000672681.1
ENST00000370188.7 |
PLPPR5
|
phospholipid phosphatase related 5 |
| chr19_-_38388020 | 0.72 |
ENST00000334928.11
ENST00000587676.1 |
GGN
|
gametogenetin |
| chr2_-_219543537 | 0.69 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr3_+_35640156 | 0.69 |
ENST00000421492.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr14_+_75069577 | 0.67 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr22_+_19718390 | 0.67 |
ENST00000383045.7
ENST00000438754.6 |
SEPTIN5
|
septin 5 |
| chr1_+_77283071 | 0.67 |
ENST00000478407.1
|
AK5
|
adenylate kinase 5 |
| chr2_-_219387881 | 0.67 |
ENST00000322176.11
ENST00000273075.9 |
DNPEP
|
aspartyl aminopeptidase |
| chr6_-_110179623 | 0.66 |
ENST00000265601.7
ENST00000447287.5 ENST00000392589.6 ENST00000444391.5 |
WASF1
|
WASP family member 1 |
| chr1_+_167936818 | 0.66 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr2_+_178194460 | 0.64 |
ENST00000392505.6
ENST00000359685.7 ENST00000357080.8 ENST00000190611.9 ENST00000409045.7 |
OSBPL6
|
oxysterol binding protein like 6 |
| chr6_-_6007511 | 0.63 |
ENST00000616243.1
|
NRN1
|
neuritin 1 |
| chr9_+_127612222 | 0.62 |
ENST00000637953.1
ENST00000636962.2 |
STXBP1
|
syntaxin binding protein 1 |
| chr6_-_109691160 | 0.62 |
ENST00000424296.7
ENST00000368948.6 ENST00000285397.9 |
AK9
|
adenylate kinase 9 |
| chr17_-_10114546 | 0.62 |
ENST00000323816.8
|
GAS7
|
growth arrest specific 7 |
| chr1_-_54887161 | 0.61 |
ENST00000535035.6
ENST00000371269.9 ENST00000436604.2 |
DHCR24
|
24-dehydrocholesterol reductase |
| chr1_-_118988301 | 0.60 |
ENST00000369429.5
|
TBX15
|
T-box transcription factor 15 |
| chr15_-_92655712 | 0.59 |
ENST00000327355.6
|
FAM174B
|
family with sequence similarity 174 member B |
| chr6_-_110179702 | 0.58 |
ENST00000392587.6
|
WASF1
|
WASP family member 1 |
| chr7_-_8262533 | 0.58 |
ENST00000447326.5
ENST00000406470.6 ENST00000407906.5 |
ICA1
|
islet cell autoantigen 1 |
| chr13_-_46897021 | 0.58 |
ENST00000542664.4
ENST00000543956.5 |
HTR2A
|
5-hydroxytryptamine receptor 2A |
| chr22_+_50600783 | 0.57 |
ENST00000329492.6
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr7_-_8262052 | 0.57 |
ENST00000396675.7
ENST00000430867.5 ENST00000402384.8 ENST00000265577.11 |
ICA1
|
islet cell autoantigen 1 |
| chr16_-_10182394 | 0.56 |
ENST00000330684.4
|
GRIN2A
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chrX_+_47078380 | 0.56 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chr2_-_72825982 | 0.56 |
ENST00000634650.1
ENST00000272427.11 ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr1_-_99004812 | 0.56 |
ENST00000263177.5
|
PLPPR5
|
phospholipid phosphatase related 5 |
| chr9_+_127612257 | 0.55 |
ENST00000637173.2
ENST00000630492.2 ENST00000627871.2 ENST00000373302.8 ENST00000373299.5 ENST00000650920.1 ENST00000476182.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr14_+_41608344 | 0.55 |
ENST00000554120.5
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr14_-_94452766 | 0.55 |
ENST00000334708.4
|
SERPINA11
|
serpin family A member 11 |
| chr3_+_42685535 | 0.54 |
ENST00000287777.5
|
KLHL40
|
kelch like family member 40 |
| chr15_+_78340344 | 0.54 |
ENST00000299529.7
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr3_-_51968387 | 0.54 |
ENST00000490063.5
ENST00000468324.5 ENST00000497653.5 ENST00000484633.5 |
PCBP4
|
poly(rC) binding protein 4 |
| chr3_-_50567711 | 0.54 |
ENST00000357203.8
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr9_+_70043840 | 0.53 |
ENST00000377182.5
|
MAMDC2
|
MAM domain containing 2 |
| chr9_-_69014090 | 0.52 |
ENST00000377276.5
|
PRKACG
|
protein kinase cAMP-activated catalytic subunit gamma |
| chr17_-_29140373 | 0.50 |
ENST00000533420.3
|
MYO18A
|
myosin XVIIIA |
| chr16_+_24845841 | 0.50 |
ENST00000347898.7
ENST00000545376.5 ENST00000449109.6 ENST00000424767.6 |
SLC5A11
|
solute carrier family 5 member 11 |
| chrX_+_47078434 | 0.50 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr1_+_213987929 | 0.49 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr5_-_150290093 | 0.49 |
ENST00000672479.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr17_-_42979993 | 0.49 |
ENST00000409103.5
ENST00000360221.8 ENST00000454303.1 ENST00000591916.7 ENST00000451885.3 |
PTGES3L-AARSD1
PTGES3L
|
PTGES3L-AARSD1 readthrough prostaglandin E synthase 3 like |
| chrX_+_47078330 | 0.48 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chr3_-_138834867 | 0.48 |
ENST00000674063.1
|
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| chr11_-_118152879 | 0.48 |
ENST00000529878.1
|
SCN4B
|
sodium voltage-gated channel beta subunit 4 |
| chr1_+_75724431 | 0.47 |
ENST00000541113.6
ENST00000680805.1 |
ACADM
|
acyl-CoA dehydrogenase medium chain |
| chr12_+_57611420 | 0.47 |
ENST00000286494.9
|
ARHGEF25
|
Rho guanine nucleotide exchange factor 25 |
| chr2_-_219543793 | 0.46 |
ENST00000243776.11
|
CHPF
|
chondroitin polymerizing factor |
| chr17_+_29929894 | 0.45 |
ENST00000536908.6
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr17_-_7219813 | 0.45 |
ENST00000399510.8
ENST00000648172.8 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr2_-_219387784 | 0.45 |
ENST00000520694.6
|
DNPEP
|
aspartyl aminopeptidase |
| chr15_-_41332487 | 0.44 |
ENST00000560640.1
ENST00000220514.8 |
OIP5
|
Opa interacting protein 5 |
| chr2_-_219387808 | 0.44 |
ENST00000521459.5
|
DNPEP
|
aspartyl aminopeptidase |
| chr1_-_205455954 | 0.43 |
ENST00000495594.2
ENST00000629624.2 |
LEMD1
BLACAT1
|
LEM domain containing 1 bladder cancer associated transcript 1 |
| chr17_-_58488375 | 0.42 |
ENST00000323777.8
|
HSF5
|
heat shock transcription factor 5 |
| chr2_-_219571241 | 0.42 |
ENST00000373876.5
ENST00000603926.5 ENST00000373873.8 ENST00000289656.3 |
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr11_-_83682385 | 0.41 |
ENST00000426717.6
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr22_-_29315656 | 0.41 |
ENST00000401450.3
ENST00000216101.7 |
RASL10A
|
RAS like family 10 member A |
| chr4_-_52038260 | 0.41 |
ENST00000381431.10
|
SGCB
|
sarcoglycan beta |
| chr15_+_74174403 | 0.40 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine rich repeat |
| chr22_+_37986156 | 0.40 |
ENST00000333418.4
ENST00000427034.1 |
POLR2F
|
RNA polymerase II, I and III subunit F |
| chr17_-_44830774 | 0.40 |
ENST00000590758.3
ENST00000591424.5 |
GJC1
|
gap junction protein gamma 1 |
| chr1_-_167937037 | 0.40 |
ENST00000271373.9
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr5_-_138543198 | 0.39 |
ENST00000507939.5
ENST00000572514.5 ENST00000499810.6 ENST00000360541.10 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr14_-_105168753 | 0.39 |
ENST00000331782.8
ENST00000347004.2 |
JAG2
|
jagged canonical Notch ligand 2 |
| chr5_+_114362286 | 0.39 |
ENST00000610748.4
ENST00000264773.7 |
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr4_-_76421121 | 0.39 |
ENST00000682701.1
|
CCDC158
|
coiled-coil domain containing 158 |
| chr16_+_24845987 | 0.39 |
ENST00000565769.5
ENST00000569520.5 |
SLC5A11
|
solute carrier family 5 member 11 |
| chr19_-_51368979 | 0.39 |
ENST00000601435.1
ENST00000291715.5 |
CLDND2
|
claudin domain containing 2 |
| chr18_-_31684504 | 0.38 |
ENST00000383131.3
ENST00000237019.11 ENST00000306851.10 |
B4GALT6
|
beta-1,4-galactosyltransferase 6 |
| chr1_+_43979877 | 0.38 |
ENST00000356836.10
ENST00000309519.8 |
B4GALT2
|
beta-1,4-galactosyltransferase 2 |
| chr1_+_14924100 | 0.38 |
ENST00000361144.9
|
KAZN
|
kazrin, periplakin interacting protein |
| chr19_+_17470474 | 0.38 |
ENST00000598424.5
ENST00000252595.12 |
SLC27A1
|
solute carrier family 27 member 1 |
| chr22_+_20967243 | 0.37 |
ENST00000683034.1
ENST00000440238.4 ENST00000405089.5 |
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr2_-_33599269 | 0.37 |
ENST00000431950.1
ENST00000403368.1 ENST00000238823.13 |
FAM98A
|
family with sequence similarity 98 member A |
| chr14_-_59630806 | 0.37 |
ENST00000342503.8
|
RTN1
|
reticulon 1 |
| chr4_-_152352800 | 0.37 |
ENST00000393956.9
|
FBXW7
|
F-box and WD repeat domain containing 7 |
| chr7_-_8262668 | 0.36 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1 |
| chrX_-_63785510 | 0.36 |
ENST00000437457.6
ENST00000374878.5 ENST00000623517.3 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr3_-_50567646 | 0.36 |
ENST00000426034.5
ENST00000441239.5 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr17_+_29593118 | 0.35 |
ENST00000394859.8
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr19_-_38387992 | 0.35 |
ENST00000586599.1
|
GGN
|
gametogenetin |
| chr7_+_148698857 | 0.34 |
ENST00000663835.1
ENST00000655324.1 ENST00000662132.1 ENST00000666124.1 ENST00000325222.9 ENST00000660240.1 |
CUL1
|
cullin 1 |
| chr20_+_43514426 | 0.34 |
ENST00000422861.3
|
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1 |
| chrX_+_43654888 | 0.34 |
ENST00000542639.5
|
MAOA
|
monoamine oxidase A |
| chr22_-_20320009 | 0.34 |
ENST00000405465.3
ENST00000248879.8 |
DGCR6L
|
DiGeorge syndrome critical region gene 6 like |
| chr12_+_14973020 | 0.34 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H |
| chrX_-_27981449 | 0.34 |
ENST00000441525.4
|
DCAF8L1
|
DDB1 and CUL4 associated factor 8 like 1 |
| chrX_+_12138426 | 0.34 |
ENST00000380682.5
ENST00000675598.1 |
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr18_+_58341038 | 0.34 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr20_+_346769 | 0.33 |
ENST00000609179.5
|
NRSN2
|
neurensin 2 |
| chr3_-_138834752 | 0.33 |
ENST00000477593.5
ENST00000483968.5 |
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| chr11_+_66857056 | 0.33 |
ENST00000309602.5
ENST00000393952.3 |
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr3_+_23945271 | 0.33 |
ENST00000312521.9
|
NR1D2
|
nuclear receptor subfamily 1 group D member 2 |
| chrX_+_153781001 | 0.33 |
ENST00000393786.7
|
SRPK3
|
SRSF protein kinase 3 |
| chr2_-_44361555 | 0.32 |
ENST00000409957.5
|
PREPL
|
prolyl endopeptidase like |
| chr19_-_45785659 | 0.32 |
ENST00000537879.1
ENST00000596586.5 ENST00000595946.1 |
DMWD
ENSG00000268434.5
|
DM1 locus, WD repeat containing novel protein |
| chr11_-_62921807 | 0.32 |
ENST00000543973.1
|
CHRM1
|
cholinergic receptor muscarinic 1 |
| chr17_-_29005913 | 0.32 |
ENST00000442608.7
ENST00000317338.17 ENST00000335960.10 |
SEZ6
|
seizure related 6 homolog |
| chr1_-_67134964 | 0.32 |
ENST00000684719.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr12_-_16605939 | 0.32 |
ENST00000541295.5
ENST00000535535.5 |
LMO3
|
LIM domain only 3 |
| chr20_+_43514492 | 0.32 |
ENST00000373135.8
ENST00000373134.5 |
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1 |
| chr15_-_29968864 | 0.32 |
ENST00000356107.11
|
TJP1
|
tight junction protein 1 |
| chr3_+_173396107 | 0.32 |
ENST00000423427.1
|
NLGN1
|
neuroligin 1 |
| chr2_+_219544002 | 0.31 |
ENST00000421791.1
ENST00000373883.4 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr12_-_16606102 | 0.31 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3 |
| chr19_-_14206168 | 0.30 |
ENST00000361434.7
ENST00000340736.10 |
ADGRL1
|
adhesion G protein-coupled receptor L1 |
| chr22_-_29061831 | 0.30 |
ENST00000216071.5
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr2_-_219571529 | 0.30 |
ENST00000404537.6
|
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr9_+_15422704 | 0.30 |
ENST00000380821.7
ENST00000610884.4 ENST00000421710.5 |
SNAPC3
|
small nuclear RNA activating complex polypeptide 3 |
| chr15_+_73052571 | 0.30 |
ENST00000560262.5
ENST00000558964.5 |
NEO1
|
neogenin 1 |
| chrX_-_112082776 | 0.30 |
ENST00000262839.3
|
TRPC5
|
transient receptor potential cation channel subfamily C member 5 |
| chr19_-_48513919 | 0.29 |
ENST00000650440.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr5_+_114362043 | 0.29 |
ENST00000673685.1
|
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr2_-_44361485 | 0.29 |
ENST00000438314.1
ENST00000409411.6 ENST00000409936.5 |
PREPL
|
prolyl endopeptidase like |
| chr19_+_42325550 | 0.29 |
ENST00000334370.8
ENST00000378073.5 |
MEGF8
|
multiple EGF like domains 8 |
| chr6_+_46793379 | 0.29 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr17_-_8118489 | 0.29 |
ENST00000380149.6
ENST00000448843.7 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr19_-_12775513 | 0.28 |
ENST00000397668.8
ENST00000587178.1 ENST00000264827.9 |
HOOK2
|
hook microtubule tethering protein 2 |
| chr18_+_48539988 | 0.28 |
ENST00000587752.5
ENST00000588345.1 |
CTIF
|
cap binding complex dependent translation initiation factor |
| chr4_-_152779710 | 0.28 |
ENST00000304337.3
|
TIGD4
|
tigger transposable element derived 4 |
| chr10_-_97771954 | 0.28 |
ENST00000266066.4
|
SFRP5
|
secreted frizzled related protein 5 |
| chr9_+_132161672 | 0.28 |
ENST00000372179.7
|
NTNG2
|
netrin G2 |
| chr6_-_40587314 | 0.28 |
ENST00000338305.7
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr19_+_46602050 | 0.28 |
ENST00000599839.5
ENST00000596362.1 |
CALM3
|
calmodulin 3 |
| chr3_-_19946970 | 0.27 |
ENST00000344838.8
|
EFHB
|
EF-hand domain family member B |
| chr9_-_124500986 | 0.27 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr1_+_109910892 | 0.27 |
ENST00000369802.7
ENST00000420111.6 |
CSF1
|
colony stimulating factor 1 |
| chr11_-_5324297 | 0.27 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2 |
| chr17_-_44830242 | 0.26 |
ENST00000592524.6
|
GJC1
|
gap junction protein gamma 1 |
| chr9_+_12695702 | 0.26 |
ENST00000381136.2
|
TYRP1
|
tyrosinase related protein 1 |
| chrX_+_129540236 | 0.26 |
ENST00000371113.9
ENST00000357121.5 |
OCRL
|
OCRL inositol polyphosphate-5-phosphatase |
| chr2_-_219387346 | 0.26 |
ENST00000430206.5
ENST00000429013.5 |
DNPEP
|
aspartyl aminopeptidase |
| chr1_+_43358968 | 0.26 |
ENST00000310955.11
|
CDC20
|
cell division cycle 20 |
| chr19_+_42325612 | 0.25 |
ENST00000251268.11
|
MEGF8
|
multiple EGF like domains 8 |
| chr9_+_132162045 | 0.24 |
ENST00000393229.4
|
NTNG2
|
netrin G2 |
| chr1_-_26900437 | 0.24 |
ENST00000361720.10
|
GPATCH3
|
G-patch domain containing 3 |
| chr19_+_45039040 | 0.24 |
ENST00000221455.8
ENST00000391953.8 ENST00000588936.5 |
CLASRP
|
CLK4 associating serine/arginine rich protein |
| chr15_+_67823557 | 0.24 |
ENST00000554240.5
|
SKOR1
|
SKI family transcriptional corepressor 1 |
| chr9_+_4490388 | 0.24 |
ENST00000262352.8
|
SLC1A1
|
solute carrier family 1 member 1 |
| chr2_+_200510003 | 0.24 |
ENST00000418045.5
|
SGO2
|
shugoshin 2 |
| chr9_-_130939068 | 0.24 |
ENST00000448616.5
ENST00000451466.1 |
FIBCD1
|
fibrinogen C domain containing 1 |
| chr8_-_92103217 | 0.24 |
ENST00000615601.4
ENST00000523629.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr1_+_14945775 | 0.24 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr17_+_6071075 | 0.24 |
ENST00000574232.5
ENST00000539421.1 |
WSCD1
|
WSC domain containing 1 |
| chr11_-_83682414 | 0.24 |
ENST00000404783.7
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr19_+_639895 | 0.23 |
ENST00000586042.6
ENST00000215530.6 |
FGF22
|
fibroblast growth factor 22 |
| chr11_+_7513768 | 0.23 |
ENST00000528947.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr12_+_56224318 | 0.23 |
ENST00000267023.9
ENST00000380198.6 ENST00000341463.5 |
NABP2
|
nucleic acid binding protein 2 |
| chr19_+_35641728 | 0.23 |
ENST00000619399.4
ENST00000379026.6 ENST00000379023.8 ENST00000402764.6 ENST00000479824.5 |
ETV2
|
ETS variant transcription factor 2 |
| chr16_-_57946602 | 0.23 |
ENST00000564654.1
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr9_-_4741176 | 0.23 |
ENST00000381809.8
|
AK3
|
adenylate kinase 3 |
| chr20_+_1135217 | 0.22 |
ENST00000381898.5
|
PSMF1
|
proteasome inhibitor subunit 1 |
| chr15_-_88467353 | 0.22 |
ENST00000312475.5
ENST00000558531.1 |
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr7_+_148698517 | 0.22 |
ENST00000665936.1
ENST00000663044.1 ENST00000602748.5 |
CUL1
|
cullin 1 |
| chr21_+_25639251 | 0.22 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr20_+_18288477 | 0.22 |
ENST00000377671.7
ENST00000360010.9 ENST00000628216.2 ENST00000425686.3 ENST00000434018.5 ENST00000630056.1 |
ZNF133
|
zinc finger protein 133 |
| chr11_+_117232625 | 0.21 |
ENST00000534428.5
ENST00000300650.9 |
RNF214
|
ring finger protein 214 |
| chr19_+_17871937 | 0.21 |
ENST00000222248.4
|
SLC5A5
|
solute carrier family 5 member 5 |
| chr1_+_20552515 | 0.21 |
ENST00000332947.6
|
FAM43B
|
family with sequence similarity 43 member B |
| chr21_+_25639272 | 0.21 |
ENST00000400532.5
ENST00000312957.9 |
JAM2
|
junctional adhesion molecule 2 |
| chr2_+_232406836 | 0.21 |
ENST00000295453.8
|
ALPG
|
alkaline phosphatase, germ cell |
| chr20_+_347024 | 0.21 |
ENST00000382291.7
ENST00000609504.5 ENST00000382285.7 ENST00000608467.5 |
NRSN2
|
neurensin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.4 | 1.5 | GO:1903633 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.3 | 1.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.2 | 1.6 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.2 | 0.7 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.2 | 0.6 | GO:0006186 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.2 | 0.8 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.2 | 1.2 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.2 | 0.5 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 | 0.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 4.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.5 | GO:0051793 | medium-chain fatty acid catabolic process(GO:0051793) |
| 0.1 | 1.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 2.1 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 0.5 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.7 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.9 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.6 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.6 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.6 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.3 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.3 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.1 | 0.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0098942 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.1 | 0.2 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.4 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 1.7 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.9 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.3 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.6 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.9 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 1.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.4 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0034553 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 1.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.5 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0060174 | limb bud formation(GO:0060174) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 2.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.7 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.2 | GO:0030892 | mitotic cohesin complex(GO:0030892) |
| 0.1 | 1.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 2.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 4.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:1990696 | stereocilia ankle link(GO:0002141) USH2 complex(GO:1990696) |
| 0.0 | 1.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.3 | 0.8 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.2 | 1.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.2 | 1.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 2.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 2.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.6 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 5.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 1.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.8 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 3.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.4 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |