Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for HOXD10
Z-value: 1.66
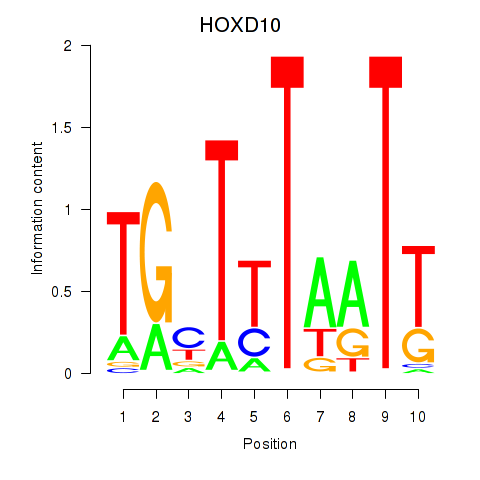
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.6 | HOXD10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg38_v1_chr2_+_176116768_176116794 | -0.06 | 7.6e-01 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_186717348 | 5.71 |
ENST00000447445.1
ENST00000287611.8 ENST00000644859.2 |
KNG1
|
kininogen 1 |
| chr4_+_154563003 | 4.04 |
ENST00000302068.9
ENST00000509493.1 |
FGB
|
fibrinogen beta chain |
| chr2_+_167248638 | 4.04 |
ENST00000295237.10
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr1_-_237945275 | 3.96 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2 like 11 |
| chr9_-_101435760 | 2.93 |
ENST00000647789.2
ENST00000616752.1 |
ALDOB
|
aldolase, fructose-bisphosphate B |
| chr4_-_71784046 | 2.52 |
ENST00000513476.5
ENST00000273951.13 |
GC
|
GC vitamin D binding protein |
| chr12_+_20810698 | 2.52 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr4_+_119135825 | 2.51 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2 |
| chr3_+_109136707 | 1.90 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr6_+_160702238 | 1.85 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_-_130222813 | 1.75 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr1_+_207104226 | 1.72 |
ENST00000367070.8
|
C4BPA
|
complement component 4 binding protein alpha |
| chr4_+_99574812 | 1.71 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr12_-_9999176 | 1.69 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr4_-_76023489 | 1.64 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr17_+_19533818 | 1.61 |
ENST00000436810.6
ENST00000270570.8 ENST00000575023.5 ENST00000395585.5 |
SLC47A1
|
solute carrier family 47 member 1 |
| chr19_-_47886308 | 1.59 |
ENST00000222002.4
|
SULT2A1
|
sulfotransferase family 2A member 1 |
| chr6_-_132734692 | 1.55 |
ENST00000509351.5
ENST00000417437.6 ENST00000423615.6 ENST00000427187.6 ENST00000414302.7 ENST00000367927.9 ENST00000450865.2 |
VNN3
|
vanin 3 |
| chr5_+_157180816 | 1.50 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr14_+_22304051 | 1.48 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr4_+_40196907 | 1.45 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr1_-_89270751 | 1.38 |
ENST00000370459.7
|
GBP5
|
guanylate binding protein 5 |
| chr15_+_48206286 | 1.35 |
ENST00000396577.7
ENST00000380993.8 |
SLC12A1
|
solute carrier family 12 member 1 |
| chr13_+_75804169 | 1.35 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr5_-_135954962 | 1.30 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr4_+_186227501 | 1.30 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr4_-_23881282 | 1.29 |
ENST00000613098.4
|
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr7_+_138076453 | 1.29 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr11_+_22668101 | 1.28 |
ENST00000630668.2
ENST00000278187.7 |
GAS2
|
growth arrest specific 2 |
| chr7_+_116811048 | 1.27 |
ENST00000464223.5
ENST00000484325.1 |
CAPZA2
|
capping actin protein of muscle Z-line subunit alpha 2 |
| chr6_-_32668368 | 1.20 |
ENST00000399084.5
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr7_-_113919000 | 1.19 |
ENST00000284601.4
|
PPP1R3A
|
protein phosphatase 1 regulatory subunit 3A |
| chr1_+_207104287 | 1.18 |
ENST00000421786.5
|
C4BPA
|
complement component 4 binding protein alpha |
| chr20_-_36646146 | 1.15 |
ENST00000262866.9
|
SLA2
|
Src like adaptor 2 |
| chr1_-_111200633 | 1.13 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr11_-_93537903 | 1.12 |
ENST00000527149.5
|
SMCO4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr1_+_109466527 | 1.11 |
ENST00000369872.4
|
SYPL2
|
synaptophysin like 2 |
| chr16_+_21612637 | 1.09 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr1_+_198638457 | 1.09 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr10_-_103153609 | 1.09 |
ENST00000675985.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr5_-_56233287 | 1.09 |
ENST00000513241.2
ENST00000341048.9 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr13_-_46211841 | 1.06 |
ENST00000442275.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr11_-_13495984 | 1.05 |
ENST00000282091.6
|
PTH
|
parathyroid hormone |
| chr3_-_190862688 | 1.05 |
ENST00000442080.6
|
GMNC
|
geminin coiled-coil domain containing |
| chr7_+_138076422 | 1.05 |
ENST00000432161.5
|
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr11_-_14972273 | 1.04 |
ENST00000396372.2
ENST00000361010.7 ENST00000331587.9 |
CALCA
|
calcitonin related polypeptide alpha |
| chr16_-_84617547 | 1.04 |
ENST00000567786.2
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr8_-_65789084 | 1.03 |
ENST00000379419.8
|
PDE7A
|
phosphodiesterase 7A |
| chr6_-_32530268 | 1.02 |
ENST00000374975.4
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr1_-_229434086 | 0.99 |
ENST00000366684.7
ENST00000684723.1 ENST00000366683.4 |
ACTA1
|
actin alpha 1, skeletal muscle |
| chr2_+_90234809 | 0.96 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr6_+_10585748 | 0.94 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr18_+_3252208 | 0.93 |
ENST00000578562.6
|
MYL12A
|
myosin light chain 12A |
| chr6_-_134318097 | 0.93 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr13_+_75804221 | 0.93 |
ENST00000489941.6
ENST00000525373.5 |
LMO7
|
LIM domain 7 |
| chr4_+_85827745 | 0.92 |
ENST00000509300.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_40197023 | 0.92 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr11_-_13496018 | 0.91 |
ENST00000529816.1
|
PTH
|
parathyroid hormone |
| chr11_+_7088991 | 0.88 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr11_+_60455839 | 0.88 |
ENST00000532491.5
ENST00000532073.5 ENST00000345732.9 ENST00000534668.6 ENST00000528313.1 ENST00000533306.6 ENST00000674194.1 |
MS4A1
|
membrane spanning 4-domains A1 |
| chr15_+_58410543 | 0.88 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr11_-_26572102 | 0.87 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr13_-_75366973 | 0.85 |
ENST00000648194.1
|
TBC1D4
|
TBC1 domain family member 4 |
| chr4_-_76007501 | 0.84 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr17_-_10549612 | 0.84 |
ENST00000532183.6
ENST00000397183.6 ENST00000420805.1 |
MYH2
|
myosin heavy chain 2 |
| chr2_+_233681877 | 0.83 |
ENST00000373426.4
|
UGT1A7
|
UDP glucuronosyltransferase family 1 member A7 |
| chr3_+_171843337 | 0.82 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr14_-_22823386 | 0.82 |
ENST00000554741.5
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr1_-_107688492 | 0.81 |
ENST00000415432.6
|
VAV3
|
vav guanine nucleotide exchange factor 3 |
| chr6_-_132763424 | 0.81 |
ENST00000532012.1
ENST00000525270.5 ENST00000530536.5 ENST00000524919.5 |
VNN2
|
vanin 2 |
| chr17_-_10549652 | 0.81 |
ENST00000245503.10
|
MYH2
|
myosin heavy chain 2 |
| chr13_+_75804270 | 0.80 |
ENST00000447038.5
|
LMO7
|
LIM domain 7 |
| chr14_+_21918161 | 0.80 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr7_-_122702912 | 0.79 |
ENST00000447240.1
ENST00000434824.2 |
RNF148
|
ring finger protein 148 |
| chr17_-_10549694 | 0.78 |
ENST00000622564.4
|
MYH2
|
myosin heavy chain 2 |
| chr15_-_55270280 | 0.78 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_-_73408313 | 0.78 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr3_+_108822759 | 0.77 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chrX_+_37780049 | 0.77 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr3_+_46370854 | 0.76 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr10_+_94683722 | 0.76 |
ENST00000285979.11
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr12_+_8913875 | 0.76 |
ENST00000538657.5
|
PHC1
|
polyhomeotic homolog 1 |
| chr3_+_101827982 | 0.76 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr18_-_28036585 | 0.75 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr1_+_198638968 | 0.75 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr4_+_99511008 | 0.75 |
ENST00000514652.5
ENST00000326581.9 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr15_+_76336755 | 0.75 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chrX_+_12906612 | 0.75 |
ENST00000218032.7
|
TLR8
|
toll like receptor 8 |
| chr7_+_142560423 | 0.75 |
ENST00000620569.1
|
TRBV12-3
|
T cell receptor beta variable 12-3 |
| chr8_+_54616057 | 0.74 |
ENST00000637698.1
|
RP1
|
RP1 axonemal microtubule associated |
| chr1_+_40247926 | 0.74 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chrX_-_15315615 | 0.74 |
ENST00000380470.7
ENST00000480796.6 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr1_-_92486916 | 0.74 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr2_+_87338511 | 0.73 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr7_+_77840122 | 0.73 |
ENST00000450574.5
ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr11_-_59212869 | 0.73 |
ENST00000361050.4
|
MPEG1
|
macrophage expressed 1 |
| chr18_+_3252267 | 0.72 |
ENST00000536605.1
ENST00000580887.5 |
MYL12A
|
myosin light chain 12A |
| chr20_+_59163810 | 0.72 |
ENST00000371030.4
|
ZNF831
|
zinc finger protein 831 |
| chr12_-_13095798 | 0.72 |
ENST00000396302.7
|
GSG1
|
germ cell associated 1 |
| chr6_-_25830557 | 0.71 |
ENST00000468082.1
|
SLC17A1
|
solute carrier family 17 member 1 |
| chr12_+_18738102 | 0.71 |
ENST00000317658.5
|
CAPZA3
|
capping actin protein of muscle Z-line subunit alpha 3 |
| chr9_-_101383558 | 0.70 |
ENST00000674556.1
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase |
| chr15_+_42274185 | 0.70 |
ENST00000440615.6
ENST00000318010.12 |
GANC
|
glucosidase alpha, neutral C |
| chr3_-_194398354 | 0.70 |
ENST00000401815.1
|
GP5
|
glycoprotein V platelet |
| chr10_+_94683771 | 0.69 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr6_-_25042003 | 0.69 |
ENST00000510784.8
|
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr1_+_198638723 | 0.69 |
ENST00000643513.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr3_+_108822778 | 0.68 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr12_-_13095628 | 0.67 |
ENST00000457134.6
ENST00000537302.5 |
GSG1
|
germ cell associated 1 |
| chr6_+_25754699 | 0.66 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr10_+_94762673 | 0.66 |
ENST00000480405.2
ENST00000371321.9 |
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19 |
| chr11_-_18236795 | 0.66 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr12_+_21372899 | 0.65 |
ENST00000240652.8
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr12_-_118359105 | 0.65 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr12_+_55362975 | 0.65 |
ENST00000641576.1
|
OR6C75
|
olfactory receptor family 6 subfamily C member 75 |
| chr4_-_40475749 | 0.65 |
ENST00000507180.5
|
RBM47
|
RNA binding motif protein 47 |
| chr18_+_63897152 | 0.64 |
ENST00000397996.6
ENST00000418725.1 |
SERPINB10
|
serpin family B member 10 |
| chr16_-_46763237 | 0.64 |
ENST00000536476.5
|
MYLK3
|
myosin light chain kinase 3 |
| chr18_-_2982873 | 0.64 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr12_+_28452493 | 0.64 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_236395394 | 0.63 |
ENST00000359362.6
|
EDARADD
|
EDAR associated death domain |
| chr2_+_119679184 | 0.63 |
ENST00000445518.1
ENST00000272521.7 ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr12_-_18738006 | 0.63 |
ENST00000266505.12
ENST00000543242.5 ENST00000539072.5 ENST00000541966.5 ENST00000648272.1 |
PLCZ1
|
phospholipase C zeta 1 |
| chr15_+_41256907 | 0.63 |
ENST00000560965.1
|
CHP1
|
calcineurin like EF-hand protein 1 |
| chr1_+_180230257 | 0.63 |
ENST00000263726.4
|
LHX4
|
LIM homeobox 4 |
| chr13_-_40982880 | 0.62 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr2_-_196176320 | 0.62 |
ENST00000420683.1
|
STK17B
|
serine/threonine kinase 17b |
| chr4_-_44651619 | 0.62 |
ENST00000415895.9
ENST00000332990.6 |
YIPF7
|
Yip1 domain family member 7 |
| chr16_-_47459130 | 0.62 |
ENST00000565940.6
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr3_+_63967738 | 0.62 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr1_+_192158448 | 0.62 |
ENST00000367460.4
|
RGS18
|
regulator of G protein signaling 18 |
| chr3_-_190449782 | 0.61 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr11_-_30586866 | 0.61 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr14_+_76310708 | 0.60 |
ENST00000512784.6
|
ESRRB
|
estrogen related receptor beta |
| chr3_-_108953870 | 0.60 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr6_+_26224192 | 0.59 |
ENST00000634733.1
|
H3C6
|
H3 clustered histone 6 |
| chrX_+_12906639 | 0.59 |
ENST00000311912.5
|
TLR8
|
toll like receptor 8 |
| chr15_+_94297939 | 0.59 |
ENST00000357742.9
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr1_+_241532370 | 0.59 |
ENST00000366559.9
ENST00000366557.8 |
KMO
|
kynurenine 3-monooxygenase |
| chr5_-_50441244 | 0.58 |
ENST00000303221.10
|
EMB
|
embigin |
| chr12_-_13095664 | 0.58 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr4_-_69860138 | 0.57 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chr6_-_135054848 | 0.57 |
ENST00000525067.1
ENST00000367822.9 |
HBS1L
|
HBS1 like translational GTPase |
| chr6_-_36547400 | 0.57 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr20_+_63738270 | 0.57 |
ENST00000467211.1
|
ENSG00000273047.1
|
novel transcript, LIME1-SLC2A4RG readthrough |
| chr7_-_115968302 | 0.57 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr18_+_22914115 | 0.56 |
ENST00000579124.5
ENST00000577588.5 ENST00000582354.5 ENST00000581819.5 |
RBBP8
|
RB binding protein 8, endonuclease |
| chr11_+_121102666 | 0.56 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr9_-_110337808 | 0.56 |
ENST00000374510.8
ENST00000374507.4 ENST00000423740.7 ENST00000374511.7 |
TXNDC8
|
thioredoxin domain containing 8 |
| chr1_-_197067234 | 0.55 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr4_-_83114715 | 0.55 |
ENST00000426923.2
ENST00000311507.9 ENST00000509973.5 |
PLAC8
|
placenta associated 8 |
| chr3_+_123067016 | 0.55 |
ENST00000316218.12
|
PDIA5
|
protein disulfide isomerase family A member 5 |
| chr15_+_36594868 | 0.55 |
ENST00000566807.5
ENST00000643612.1 ENST00000567389.5 ENST00000562877.5 |
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr2_+_68734861 | 0.55 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr5_+_122129533 | 0.54 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr4_-_67963441 | 0.54 |
ENST00000508048.6
|
TMPRSS11A
|
transmembrane serine protease 11A |
| chr7_+_90469634 | 0.54 |
ENST00000509356.2
|
PTTG1IP2
|
PTTG1IP family member 2 |
| chr1_+_61952283 | 0.53 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr7_+_92057602 | 0.53 |
ENST00000491695.2
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr10_-_75109085 | 0.53 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chrX_-_50200988 | 0.53 |
ENST00000358526.7
|
AKAP4
|
A-kinase anchoring protein 4 |
| chr12_+_53994874 | 0.53 |
ENST00000509328.1
|
HOXC6
|
homeobox C6 |
| chr2_-_89085787 | 0.52 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr6_-_134317905 | 0.52 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_49964160 | 0.52 |
ENST00000322066.4
|
DEFB114
|
defensin beta 114 |
| chr11_+_112176364 | 0.52 |
ENST00000526088.5
ENST00000532593.5 ENST00000531169.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr20_-_17558811 | 0.51 |
ENST00000536626.7
ENST00000377868.6 |
BFSP1
|
beaded filament structural protein 1 |
| chr7_+_106865474 | 0.51 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr7_+_138076475 | 0.51 |
ENST00000438242.1
|
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr5_-_50441350 | 0.50 |
ENST00000508934.5
|
EMB
|
embigin |
| chr17_+_42773442 | 0.50 |
ENST00000253794.7
ENST00000590339.5 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog |
| chr6_-_118710065 | 0.49 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr8_+_54616078 | 0.48 |
ENST00000220676.2
|
RP1
|
RP1 axonemal microtubule associated |
| chr11_+_112175526 | 0.48 |
ENST00000532612.5
ENST00000438022.5 |
BCO2
|
beta-carotene oxygenase 2 |
| chr1_-_75932392 | 0.48 |
ENST00000284142.7
|
ASB17
|
ankyrin repeat and SOCS box containing 17 |
| chr5_-_147906530 | 0.47 |
ENST00000318315.5
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr7_-_23347704 | 0.47 |
ENST00000619562.4
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr2_+_68734773 | 0.47 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr5_+_140875299 | 0.47 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr21_-_30216047 | 0.47 |
ENST00000399899.2
|
CLDN8
|
claudin 8 |
| chr2_+_218710931 | 0.46 |
ENST00000442769.5
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase like 4 |
| chr14_+_22096017 | 0.46 |
ENST00000390452.2
|
TRDV1
|
T cell receptor delta variable 1 |
| chr7_+_130207847 | 0.46 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr7_+_142554828 | 0.45 |
ENST00000611787.1
|
TRBV11-3
|
T cell receptor beta variable 11-3 |
| chr8_-_51809414 | 0.45 |
ENST00000356297.5
|
PXDNL
|
peroxidasin like |
| chr11_+_56027654 | 0.45 |
ENST00000641320.1
|
OR5AS1
|
olfactory receptor family 5 subfamily AS member 1 |
| chr7_-_87713287 | 0.45 |
ENST00000416177.1
ENST00000265724.8 ENST00000543898.5 |
ABCB1
|
ATP binding cassette subfamily B member 1 |
| chrX_-_21758021 | 0.44 |
ENST00000646008.1
|
SMPX
|
small muscle protein X-linked |
| chr2_+_182716227 | 0.44 |
ENST00000680258.1
ENST00000680667.1 ENST00000264065.12 ENST00000616986.5 ENST00000679884.1 |
DNAJC10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr12_-_11134644 | 0.44 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr7_+_95485934 | 0.43 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr19_-_55038256 | 0.43 |
ENST00000417454.5
ENST00000310373.7 ENST00000333884.2 |
GP6
|
glycoprotein VI platelet |
| chr1_-_202889099 | 0.43 |
ENST00000367262.4
|
RABIF
|
RAB interacting factor |
| chr4_-_122456725 | 0.43 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chrX_+_15749848 | 0.43 |
ENST00000479740.5
ENST00000454127.2 |
CA5B
|
carbonic anhydrase 5B |
| chr2_+_90172802 | 0.42 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr15_-_32454902 | 0.42 |
ENST00000562377.1
|
GOLGA8O
|
golgin A8 family member O |
| chr9_-_127755243 | 0.42 |
ENST00000629203.2
ENST00000420366.5 |
SH2D3C
|
SH2 domain containing 3C |
| chr7_-_38249572 | 0.42 |
ENST00000436911.6
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr12_+_103965835 | 0.42 |
ENST00000266775.13
ENST00000544861.5 |
TDG
|
thymine DNA glycosylase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.8 | 3.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.5 | 2.5 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.4 | 1.3 | GO:0071250 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) response to resveratrol(GO:1904638) cellular response to resveratrol(GO:1904639) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.4 | 2.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.3 | 1.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.3 | 1.0 | GO:0046247 | tetraterpenoid metabolic process(GO:0016108) carotenoid metabolic process(GO:0016116) carotene catabolic process(GO:0016121) xanthophyll metabolic process(GO:0016122) terpene catabolic process(GO:0046247) |
| 0.3 | 2.9 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.3 | 3.0 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.3 | 4.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.3 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.3 | 0.8 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.7 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 0.7 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 6.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 0.6 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 0.6 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 1.6 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 1.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.4 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 0.6 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.6 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 2.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.8 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 2.0 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.4 | GO:1903124 | regulation of thioredoxin peroxidase activity(GO:1903123) negative regulation of thioredoxin peroxidase activity(GO:1903124) negative regulation of thioredoxin peroxidase activity by peptidyl-threonine phosphorylation(GO:1903125) Wnt signalosome assembly(GO:1904887) negative regulation of peroxidase activity(GO:2000469) |
| 0.1 | 0.6 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 | 0.4 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.9 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 1.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.7 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 0.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.9 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.4 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.5 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.7 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.1 | 0.7 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 2.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.8 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 1.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.8 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.8 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.4 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.7 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 2.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:0033320 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.1 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.4 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 2.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0042779 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0035853 | chromosome passenger complex localization to spindle midzone(GO:0035853) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.6 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.5 | GO:1903961 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 4.0 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.2 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.6 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 1.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0044107 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 1.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 2.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.8 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.7 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 1.2 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.7 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 2.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 2.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1990844 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.3 | 4.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 1.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 2.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.4 | GO:0032473 | cytoplasmic side of mitochondrial outer membrane(GO:0032473) caveola neck(GO:0099400) |
| 0.1 | 2.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 2.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.3 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.1 | 0.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 12.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 3.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 7.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) translation initiation ternary complex(GO:0044207) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.6 | 2.5 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.6 | 1.8 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.6 | 2.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.6 | 2.8 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.5 | 2.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 1.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.4 | 1.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.3 | 2.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.3 | 2.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 1.6 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.3 | 1.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 0.7 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.2 | 0.6 | GO:0005139 | interleukin-7 receptor binding(GO:0005139) |
| 0.2 | 0.5 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 0.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.4 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) peroxidase inhibitor activity(GO:0036479) |
| 0.1 | 0.4 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 1.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 2.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.1 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 3.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 2.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 1.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.2 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.4 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 4.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 4.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.8 | GO:0050664 | superoxide-generating NADPH oxidase activity(GO:0016175) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 2.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.6 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 3.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 4.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 3.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 7.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 3.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 0.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 3.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 2.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 3.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |