Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
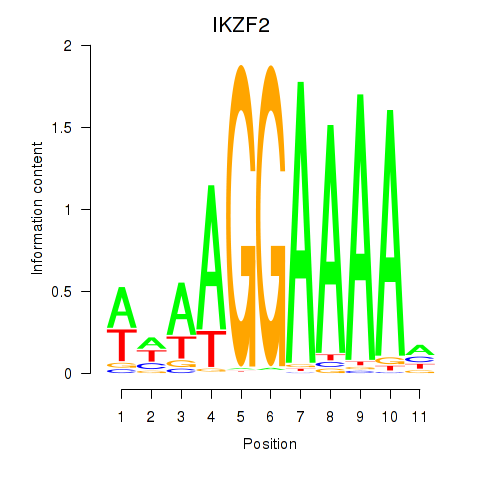
Results for IKZF2
Z-value: 2.08
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.17 | IKZF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg38_v1_chr2_-_213151590_213151619 | 0.31 | 8.4e-02 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_70666492 | 7.60 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr4_-_70666884 | 7.00 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr14_-_106235582 | 4.69 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr14_-_106117159 | 4.59 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr4_+_67558719 | 4.27 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr12_-_121802886 | 3.65 |
ENST00000545885.5
ENST00000542933.5 ENST00000428029.6 ENST00000541694.5 ENST00000536662.5 ENST00000535643.5 ENST00000541657.5 |
LINC01089
RHOF
|
long intergenic non-protein coding RNA 1089 ras homolog family member F, filopodia associated |
| chr14_-_106791226 | 3.35 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr2_+_97713568 | 3.31 |
ENST00000264972.10
|
ZAP70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr17_+_7884783 | 3.27 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr1_+_100719734 | 3.03 |
ENST00000370119.8
ENST00000294728.7 ENST00000347652.6 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr14_-_106335613 | 2.74 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr7_+_142791635 | 2.72 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr14_-_106803221 | 2.71 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr7_+_142618821 | 2.68 |
ENST00000390393.3
|
TRBV19
|
T cell receptor beta variable 19 |
| chr1_+_158831323 | 2.61 |
ENST00000368141.5
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr14_-_106557465 | 2.53 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr14_-_106211453 | 2.51 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr1_-_169630115 | 2.44 |
ENST00000263686.11
ENST00000367788.6 |
SELP
|
selectin P |
| chr20_+_59604527 | 2.41 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr5_-_111756245 | 2.41 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr5_+_35852695 | 2.38 |
ENST00000508941.5
|
IL7R
|
interleukin 7 receptor |
| chr8_+_79611036 | 2.37 |
ENST00000220876.12
ENST00000518111.5 |
STMN2
|
stathmin 2 |
| chr5_-_34043205 | 2.31 |
ENST00000382065.8
ENST00000231338.7 |
C1QTNF3
|
C1q and TNF related 3 |
| chr1_+_9651723 | 2.29 |
ENST00000377346.9
ENST00000536656.5 ENST00000628140.2 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr14_-_106658251 | 2.23 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr1_-_47190013 | 2.21 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr20_+_38805686 | 2.21 |
ENST00000299824.6
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1 regulatory subunit 16B |
| chr1_-_169630076 | 2.09 |
ENST00000367786.6
ENST00000458599.6 ENST00000367795.2 |
SELP
|
selectin P |
| chr5_+_55102635 | 2.08 |
ENST00000274306.7
|
GZMA
|
granzyme A |
| chr11_-_75306688 | 2.03 |
ENST00000532525.1
|
ARRB1
|
arrestin beta 1 |
| chr14_+_51860391 | 1.95 |
ENST00000335281.8
|
GNG2
|
G protein subunit gamma 2 |
| chr1_+_171248471 | 1.93 |
ENST00000402921.6
ENST00000617670.6 ENST00000367750.7 |
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr2_+_27078598 | 1.91 |
ENST00000380320.9
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr1_+_159009886 | 1.88 |
ENST00000340979.10
ENST00000368131.8 ENST00000295809.12 ENST00000368132.7 |
IFI16
|
interferon gamma inducible protein 16 |
| chr21_-_14546297 | 1.83 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr17_+_7888783 | 1.82 |
ENST00000330494.12
ENST00000358181.8 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr3_-_112610262 | 1.81 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr3_+_196744 | 1.79 |
ENST00000256509.7
ENST00000397491.6 |
CHL1
|
cell adhesion molecule L1 like |
| chr16_+_31355165 | 1.78 |
ENST00000562918.5
ENST00000268296.9 |
ITGAX
|
integrin subunit alpha X |
| chr13_-_99307387 | 1.78 |
ENST00000376414.5
|
GPR183
|
G protein-coupled receptor 183 |
| chr17_-_48610971 | 1.76 |
ENST00000239165.9
|
HOXB7
|
homeobox B7 |
| chr16_+_31355215 | 1.76 |
ENST00000562522.2
|
ITGAX
|
integrin subunit alpha X |
| chr19_-_41353904 | 1.71 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1 |
| chr10_+_88880236 | 1.70 |
ENST00000371926.8
|
STAMBPL1
|
STAM binding protein like 1 |
| chr3_-_183555696 | 1.70 |
ENST00000341319.8
|
KLHL6
|
kelch like family member 6 |
| chr19_-_46787278 | 1.69 |
ENST00000412532.6
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr22_+_39901075 | 1.67 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chr3_+_46354072 | 1.67 |
ENST00000445132.3
ENST00000421659.1 |
CCR2
|
C-C motif chemokine receptor 2 |
| chr14_+_64465491 | 1.65 |
ENST00000394718.4
|
AKAP5
|
A-kinase anchoring protein 5 |
| chr7_+_93921720 | 1.57 |
ENST00000248564.6
|
GNG11
|
G protein subunit gamma 11 |
| chr12_-_49187369 | 1.56 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chrX_+_136648138 | 1.56 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr1_-_160579439 | 1.56 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chr6_-_111759910 | 1.55 |
ENST00000517419.5
|
FYN
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr6_+_31615215 | 1.53 |
ENST00000337917.11
ENST00000376059.8 |
AIF1
|
allograft inflammatory factor 1 |
| chr14_+_51860632 | 1.52 |
ENST00000555472.5
ENST00000556766.5 |
GNG2
|
G protein subunit gamma 2 |
| chrX_-_19670983 | 1.51 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr8_-_107498041 | 1.51 |
ENST00000297450.7
|
ANGPT1
|
angiopoietin 1 |
| chr6_-_31878967 | 1.49 |
ENST00000414427.1
ENST00000229729.11 ENST00000375562.8 |
SLC44A4
|
solute carrier family 44 member 4 |
| chr2_-_85410336 | 1.49 |
ENST00000263867.9
ENST00000409921.5 |
CAPG
|
capping actin protein, gelsolin like |
| chr8_+_103819244 | 1.48 |
ENST00000262231.14
ENST00000507740.5 ENST00000408894.6 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_159010002 | 1.46 |
ENST00000359709.7
|
IFI16
|
interferon gamma inducible protein 16 |
| chr21_-_14546351 | 1.46 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr6_+_31670379 | 1.45 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr9_-_114930508 | 1.43 |
ENST00000223795.3
ENST00000618336.4 |
TNFSF8
|
TNF superfamily member 8 |
| chr13_-_46182136 | 1.43 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chrX_-_101407893 | 1.40 |
ENST00000676156.1
ENST00000675592.1 ENST00000674634.2 ENST00000649178.1 ENST00000218516.4 |
GLA
|
galactosidase alpha |
| chrX_+_78747705 | 1.39 |
ENST00000614823.5
ENST00000435339.3 ENST00000514744.5 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr1_+_209768597 | 1.39 |
ENST00000487271.5
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr17_-_50707855 | 1.38 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr6_+_31670167 | 1.36 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr11_+_72192126 | 1.36 |
ENST00000393676.5
|
FOLR1
|
folate receptor alpha |
| chr6_+_106086316 | 1.35 |
ENST00000369091.6
ENST00000369096.9 |
PRDM1
|
PR/SET domain 1 |
| chr19_+_18386150 | 1.34 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr1_-_157700738 | 1.32 |
ENST00000368186.9
ENST00000496769.1 ENST00000368184.8 |
FCRL3
|
Fc receptor like 3 |
| chr17_-_48185179 | 1.30 |
ENST00000579336.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr1_-_68232539 | 1.30 |
ENST00000370976.7
ENST00000354777.6 |
WLS
|
Wnt ligand secretion mediator |
| chr4_-_113979635 | 1.27 |
ENST00000315366.8
|
ARSJ
|
arylsulfatase family member J |
| chr1_-_155978524 | 1.26 |
ENST00000361247.9
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr6_+_116399395 | 1.25 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr1_-_68232514 | 1.25 |
ENST00000262348.9
ENST00000370973.2 ENST00000370971.1 |
WLS
|
Wnt ligand secretion mediator |
| chr13_+_57631735 | 1.24 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr4_-_108167784 | 1.23 |
ENST00000438313.6
|
LEF1
|
lymphoid enhancer binding factor 1 |
| chr7_-_111784448 | 1.23 |
ENST00000450156.6
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr1_+_43147890 | 1.22 |
ENST00000335282.5
ENST00000409396.5 |
FAM183A
|
family with sequence similarity 183 member A |
| chrX_-_133985449 | 1.22 |
ENST00000631057.2
|
GPC3
|
glypican 3 |
| chr19_-_42442938 | 1.21 |
ENST00000601181.6
|
CXCL17
|
C-X-C motif chemokine ligand 17 |
| chr11_+_61755372 | 1.19 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chr1_+_206205909 | 1.19 |
ENST00000624873.3
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_-_33869816 | 1.19 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chr2_+_33436304 | 1.17 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr10_+_113709261 | 1.17 |
ENST00000672138.1
ENST00000452490.3 |
CASP7
|
caspase 7 |
| chrX_+_136648214 | 1.17 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr2_+_203936755 | 1.17 |
ENST00000316386.11
ENST00000435193.1 |
ICOS
|
inducible T cell costimulator |
| chr6_-_41747390 | 1.16 |
ENST00000356667.8
ENST00000373025.7 ENST00000425343.6 |
PGC
|
progastricsin |
| chrX_+_56729231 | 1.16 |
ENST00000637096.1
ENST00000374922.9 ENST00000423617.2 |
NBDY
|
negative regulator of P-body association |
| chr7_-_111784406 | 1.15 |
ENST00000664131.1
ENST00000437129.5 |
DOCK4
|
dedicator of cytokinesis 4 |
| chrX_+_103776831 | 1.13 |
ENST00000621218.5
ENST00000619236.1 |
PLP1
|
proteolipid protein 1 |
| chr12_-_50283472 | 1.13 |
ENST00000551691.5
ENST00000394943.7 ENST00000341247.8 |
LIMA1
|
LIM domain and actin binding 1 |
| chr8_-_133102477 | 1.13 |
ENST00000522119.5
ENST00000523610.5 ENST00000338087.10 ENST00000521302.5 ENST00000519558.5 ENST00000519747.5 ENST00000517648.5 |
SLA
|
Src like adaptor |
| chr1_+_206205626 | 1.12 |
ENST00000419187.6
ENST00000605610.5 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_-_161371501 | 1.12 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase small subunit B |
| chr1_-_145093246 | 1.10 |
ENST00000619678.1
|
SRGAP2B
|
SLIT-ROBO Rho GTPase activating protein 2B |
| chr6_-_159044980 | 1.09 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr6_-_131063233 | 1.09 |
ENST00000392427.7
ENST00000337057.8 ENST00000525271.5 ENST00000527411.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr6_+_32637419 | 1.09 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr6_+_26365159 | 1.08 |
ENST00000532865.5
ENST00000396934.7 ENST00000508906.6 ENST00000530653.5 ENST00000527417.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chrX_+_103776493 | 1.08 |
ENST00000433491.5
ENST00000612423.4 ENST00000443502.5 |
PLP1
|
proteolipid protein 1 |
| chr15_+_81000913 | 1.08 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr6_+_106098933 | 1.07 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr2_+_188974364 | 1.07 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr12_+_121132869 | 1.07 |
ENST00000328963.10
|
P2RX7
|
purinergic receptor P2X 7 |
| chr12_-_123436664 | 1.06 |
ENST00000280571.10
|
RILPL2
|
Rab interacting lysosomal protein like 2 |
| chr12_-_79935069 | 1.06 |
ENST00000450142.7
|
PPP1R12A
|
protein phosphatase 1 regulatory subunit 12A |
| chr9_-_136439796 | 1.05 |
ENST00000676019.1
ENST00000371712.4 |
INPP5E
|
inositol polyphosphate-5-phosphatase E |
| chr13_-_29586788 | 1.05 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 member 1 |
| chrX_+_123963164 | 1.05 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr14_+_22007503 | 1.05 |
ENST00000390447.3
|
TRAV19
|
T cell receptor alpha variable 19 |
| chr2_-_133568393 | 1.05 |
ENST00000317721.10
ENST00000405974.7 ENST00000409261.6 ENST00000409213.5 |
NCKAP5
|
NCK associated protein 5 |
| chr14_+_21642856 | 1.04 |
ENST00000390423.2
|
TRAV1-2
|
T cell receptor alpha variable 1-2 |
| chr17_-_7217206 | 1.04 |
ENST00000447163.6
ENST00000647975.1 |
DLG4
|
discs large MAGUK scaffold protein 4 |
| chr6_-_159045104 | 1.04 |
ENST00000326965.7
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr8_+_79611727 | 1.04 |
ENST00000518491.1
|
STMN2
|
stathmin 2 |
| chr7_+_94394886 | 1.04 |
ENST00000297268.11
ENST00000620463.1 |
COL1A2
|
collagen type I alpha 2 chain |
| chr6_+_32637396 | 1.03 |
ENST00000395363.5
ENST00000496318.5 ENST00000343139.11 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr10_+_30434021 | 1.02 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_-_155978427 | 1.00 |
ENST00000313667.8
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr9_+_2159850 | 1.00 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr18_-_26657401 | 1.00 |
ENST00000580191.5
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr6_-_131063272 | 1.00 |
ENST00000445890.6
ENST00000368128.6 ENST00000628542.2 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr4_-_87391149 | 0.99 |
ENST00000507286.1
ENST00000358290.9 |
HSD17B11
|
hydroxysteroid 17-beta dehydrogenase 11 |
| chr8_-_23425307 | 0.98 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase like 2 |
| chr16_+_81238682 | 0.98 |
ENST00000258168.7
ENST00000564552.1 |
BCO1
|
beta-carotene oxygenase 1 |
| chr10_+_117543567 | 0.98 |
ENST00000616794.1
|
EMX2
|
empty spiracles homeobox 2 |
| chr15_+_96333111 | 0.97 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr5_+_58491451 | 0.96 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr5_-_151686908 | 0.96 |
ENST00000231061.9
|
SPARC
|
secreted protein acidic and cysteine rich |
| chr6_-_75202792 | 0.95 |
ENST00000416123.6
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr18_-_25351615 | 0.95 |
ENST00000580488.1
ENST00000577801.5 |
ZNF521
|
zinc finger protein 521 |
| chr6_+_167122742 | 0.94 |
ENST00000341935.9
ENST00000349984.6 |
CCR6
|
C-C motif chemokine receptor 6 |
| chr2_-_189179754 | 0.94 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr6_-_159045010 | 0.94 |
ENST00000338313.5
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr6_+_26365215 | 0.93 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr2_-_175005357 | 0.93 |
ENST00000409156.7
ENST00000444573.2 ENST00000409900.9 |
CHN1
|
chimerin 1 |
| chr5_-_140633167 | 0.93 |
ENST00000302014.11
|
CD14
|
CD14 molecule |
| chr10_+_30434176 | 0.92 |
ENST00000263056.6
ENST00000375322.2 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr8_-_71356511 | 0.91 |
ENST00000419131.6
ENST00000388743.6 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr19_+_11089446 | 0.91 |
ENST00000557933.5
ENST00000455727.6 ENST00000535915.5 ENST00000545707.5 ENST00000558518.6 ENST00000558013.5 |
LDLR
|
low density lipoprotein receptor |
| chr11_-_69013356 | 0.91 |
ENST00000441623.1
|
MRGPRF
|
MAS related GPR family member F |
| chr3_-_116445458 | 0.91 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein |
| chr9_+_2159672 | 0.90 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_103571077 | 0.89 |
ENST00000610648.1
|
AMY2B
|
amylase alpha 2B |
| chr1_+_174964750 | 0.89 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr6_+_26365176 | 0.89 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr3_-_27722699 | 0.89 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr16_+_33827140 | 0.88 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr14_+_22281097 | 0.88 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr16_-_67999468 | 0.88 |
ENST00000393847.6
ENST00000573808.1 ENST00000572624.5 |
DPEP2
|
dipeptidase 2 |
| chr6_+_21593742 | 0.88 |
ENST00000244745.4
|
SOX4
|
SRY-box transcription factor 4 |
| chr6_-_11778781 | 0.87 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr17_+_56978111 | 0.87 |
ENST00000262288.8
ENST00000572710.5 ENST00000575395.5 ENST00000631024.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr17_-_7484205 | 0.87 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr7_+_142492121 | 0.86 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6 |
| chr1_+_174964820 | 0.85 |
ENST00000485114.1
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr2_+_113406368 | 0.84 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr3_+_10026409 | 0.84 |
ENST00000287647.7
ENST00000676013.1 ENST00000675286.1 ENST00000419585.5 |
FANCD2
|
FA complementation group D2 |
| chrX_-_41665766 | 0.84 |
ENST00000643043.2
ENST00000486402.1 ENST00000646087.2 |
CASK
|
calcium/calmodulin dependent serine protein kinase |
| chr13_+_48976597 | 0.83 |
ENST00000541916.5
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr1_+_78649818 | 0.81 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr2_+_66435558 | 0.81 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1 |
| chr8_-_100952002 | 0.80 |
ENST00000521309.5
ENST00000395958.6 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
| chr15_-_88256158 | 0.80 |
ENST00000317501.7
ENST00000629765.2 ENST00000557856.5 ENST00000558676.5 |
NTRK3
|
neurotrophic receptor tyrosine kinase 3 |
| chr2_-_157325808 | 0.80 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr20_+_43945677 | 0.80 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr2_+_191678122 | 0.79 |
ENST00000425611.9
ENST00000410026.7 |
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_28042660 | 0.79 |
ENST00000407008.8
|
NLK
|
nemo like kinase |
| chr15_-_55249029 | 0.79 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_+_43692886 | 0.79 |
ENST00000434505.5
ENST00000411750.5 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr11_-_102724945 | 0.78 |
ENST00000236826.8
|
MMP8
|
matrix metallopeptidase 8 |
| chr13_-_40982880 | 0.77 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chrX_-_136780925 | 0.76 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr5_+_96875978 | 0.76 |
ENST00000510373.5
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr11_-_105023136 | 0.76 |
ENST00000526056.5
ENST00000531367.5 ENST00000456094.1 ENST00000444749.6 ENST00000393141.6 ENST00000418434.5 ENST00000260315.8 |
CASP5
|
caspase 5 |
| chrX_+_120604084 | 0.76 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr18_-_55403682 | 0.75 |
ENST00000564228.5
ENST00000630828.2 |
TCF4
|
transcription factor 4 |
| chrX_-_117985298 | 0.75 |
ENST00000469946.5
|
KLHL13
|
kelch like family member 13 |
| chr14_-_35121950 | 0.75 |
ENST00000554361.5
ENST00000261475.10 |
PPP2R3C
|
protein phosphatase 2 regulatory subunit B''gamma |
| chr6_-_131063207 | 0.75 |
ENST00000530481.5
|
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr6_-_132734692 | 0.73 |
ENST00000509351.5
ENST00000417437.6 ENST00000423615.6 ENST00000427187.6 ENST00000414302.7 ENST00000367927.9 ENST00000450865.2 |
VNN3
|
vanin 3 |
| chr6_+_148508446 | 0.73 |
ENST00000637729.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr7_+_142391884 | 0.73 |
ENST00000390363.2
|
TRBV9
|
T cell receptor beta variable 9 |
| chr1_+_111139436 | 0.72 |
ENST00000545121.5
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr10_-_7410973 | 0.72 |
ENST00000684547.1
ENST00000673876.1 ENST00000397167.6 |
SFMBT2
|
Scm like with four mbt domains 2 |
| chr3_+_41194741 | 0.72 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr7_+_30852273 | 0.72 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough |
| chr17_+_28719995 | 0.71 |
ENST00000496182.5
|
RPL23A
|
ribosomal protein L23a |
| chr21_-_10649835 | 0.70 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr1_+_111346590 | 0.70 |
ENST00000369737.4
ENST00000369738.9 |
PIFO
|
primary cilia formation |
| chr15_+_81182579 | 0.69 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr2_+_191678152 | 0.69 |
ENST00000409510.5
|
NABP1
|
nucleic acid binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 1.3 | 14.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.9 | 3.4 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.8 | 2.4 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.7 | 2.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.6 | 2.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.6 | 1.7 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.6 | 3.3 | GO:0043366 | beta selection(GO:0043366) |
| 0.5 | 1.6 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.5 | 1.6 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 0.5 | 1.5 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.5 | 1.5 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.5 | 2.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.5 | 1.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 3.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 1.7 | GO:0052552 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.4 | 1.7 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.4 | 1.2 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.4 | 1.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.3 | 2.8 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 2.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.3 | 2.3 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.3 | 0.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 | 1.2 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.3 | 1.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.3 | 22.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.0 | GO:0018277 | protein deamination(GO:0018277) |
| 0.2 | 1.0 | GO:0016119 | carotene metabolic process(GO:0016119) |
| 0.2 | 1.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 4.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 0.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 2.3 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.2 | 1.5 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.2 | 1.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.2 | 2.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 2.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.2 | 1.8 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 1.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 1.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.9 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 0.9 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 0.9 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.2 | 1.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 0.9 | GO:0038124 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 1.0 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 1.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 2.9 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 1.6 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 2.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 | 1.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.8 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 1.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.6 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 1.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 1.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 1.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 1.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 1.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.7 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 3.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 2.6 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.8 | GO:1903435 | cytotoxic T cell degranulation(GO:0043316) positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 3.1 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 3.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 1.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.5 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 3.5 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 1.6 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.7 | GO:0061324 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.4 | GO:0048687 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 3.3 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.9 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.6 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.6 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.1 | 2.9 | GO:0051023 | regulation of immunoglobulin secretion(GO:0051023) |
| 0.1 | 0.6 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.6 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 1.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.7 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 1.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.6 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.5 | GO:1900223 | positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.7 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 3.3 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.1 | 7.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 1.8 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 1.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.3 | GO:0097017 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) renal protein absorption(GO:0097017) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 1.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 1.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 1.3 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.7 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.8 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 5.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 1.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 1.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 5.1 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 1.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.7 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.4 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 1.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 1.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.6 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.9 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:2000437 | regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:2000176 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.8 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.3 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 3.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 2.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.6 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.2 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.6 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 1.0 | 3.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.4 | 22.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 1.0 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.3 | 0.9 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 4.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 1.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 2.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 4.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 1.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 5.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.1 | 2.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.6 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 1.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 6.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 3.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 3.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.1 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 2.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 3.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 3.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 2.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 4.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 6.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.9 | 2.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.8 | 2.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.7 | 2.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.6 | 4.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.6 | 1.9 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.5 | 4.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.5 | 1.5 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.4 | 1.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.2 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.3 | 1.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 22.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 1.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 1.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 2.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.7 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 2.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.6 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 1.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.6 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.2 | 1.7 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 0.7 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 0.7 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 0.9 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 3.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 1.6 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 1.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 2.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.6 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 1.0 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.9 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 1.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.9 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 2.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.1 | 1.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 5.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.3 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.2 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 1.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 1.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 8.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.0 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 7.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 3.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.0 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 2.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 2.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 7.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 3.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0005030 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 4.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 7.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 3.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 9.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 4.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 4.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 3.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 4.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 5.6 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 1.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 5.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 0.5 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 2.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 8.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 13.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 3.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 1.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |