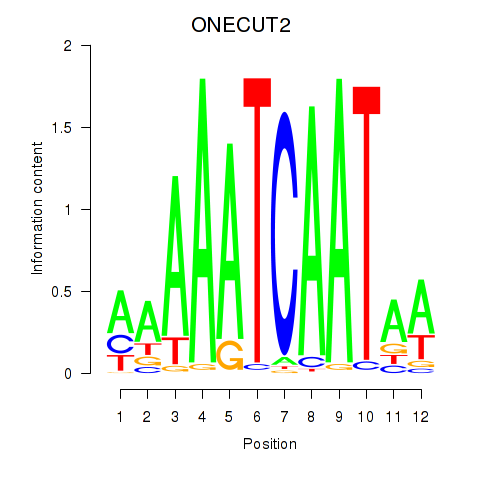
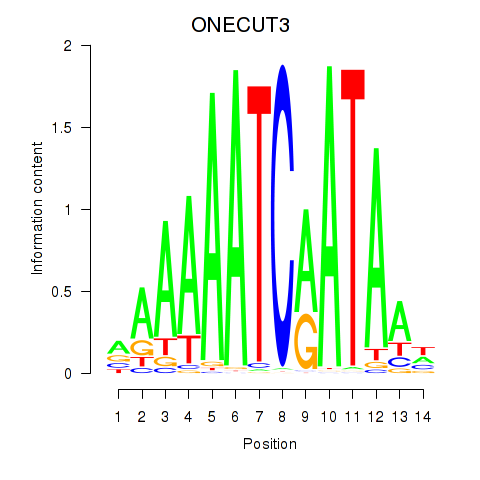
Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for ONECUT2_ONECUT3
Z-value: 1.67
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.6 | ONECUT2 |
|
ONECUT3
|
ENSG00000205922.5 | ONECUT3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT2 | hg38_v1_chr18_+_57435366_57435391 | 0.57 | 7.1e-04 | Click! |
| ONECUT3 | hg38_v1_chr19_+_1753499_1753521 | -0.06 | 7.6e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_63443076 | 7.34 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr16_+_56191476 | 6.37 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr10_+_93757857 | 5.58 |
ENST00000478763.2
ENST00000371418.9 ENST00000630184.2 ENST00000630047.2 ENST00000637689.1 |
LGI1
|
leucine rich glioma inactivated 1 |
| chr7_-_100641507 | 5.01 |
ENST00000431692.5
ENST00000223051.8 |
TFR2
|
transferrin receptor 2 |
| chr1_+_159587817 | 5.00 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chr14_-_23578756 | 4.77 |
ENST00000397118.7
ENST00000356300.9 |
JPH4
|
junctophilin 4 |
| chr13_-_35855758 | 4.30 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr10_+_93757831 | 4.08 |
ENST00000629035.2
|
LGI1
|
leucine rich glioma inactivated 1 |
| chr16_+_56191536 | 3.81 |
ENST00000569295.6
|
GNAO1
|
G protein subunit alpha o1 |
| chr13_-_35855627 | 3.55 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr3_-_187670385 | 3.41 |
ENST00000287641.4
|
SST
|
somatostatin |
| chr3_+_63443306 | 3.36 |
ENST00000472899.5
ENST00000479198.5 ENST00000460711.5 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr12_-_62192762 | 3.34 |
ENST00000416284.8
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr1_-_159714581 | 3.13 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr7_-_143362687 | 2.76 |
ENST00000409578.5
ENST00000443739.7 ENST00000409346.5 |
FAM131B
|
family with sequence similarity 131 member B |
| chr16_+_56191728 | 2.69 |
ENST00000638705.1
ENST00000262494.12 |
GNAO1
|
G protein subunit alpha o1 |
| chr11_-_790062 | 2.67 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_+_198638723 | 2.64 |
ENST00000643513.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr1_+_198638968 | 2.56 |
ENST00000348564.11
ENST00000530727.5 ENST00000442510.8 ENST00000645247.1 ENST00000367367.8 ENST00000367364.5 ENST00000413409.6 |
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr11_-_35419213 | 2.43 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr7_-_105269007 | 2.43 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr2_-_191151568 | 2.33 |
ENST00000358470.8
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr2_-_191151549 | 2.32 |
ENST00000413064.5
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr1_+_198638457 | 2.31 |
ENST00000367379.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr11_+_63369779 | 2.26 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr3_+_40100007 | 2.15 |
ENST00000539167.2
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr7_+_43112593 | 2.14 |
ENST00000453890.5
ENST00000395891.7 |
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr16_+_56328661 | 2.10 |
ENST00000562316.6
|
GNAO1
|
G protein subunit alpha o1 |
| chr20_-_1994046 | 2.04 |
ENST00000217305.3
ENST00000650874.1 |
PDYN
|
prodynorphin |
| chr17_-_66229380 | 1.96 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr17_+_46624036 | 1.93 |
ENST00000575068.5
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr12_+_56752449 | 1.93 |
ENST00000554643.5
ENST00000556650.5 ENST00000554150.5 ENST00000554155.1 |
HSD17B6
|
hydroxysteroid 17-beta dehydrogenase 6 |
| chr19_+_3880647 | 1.92 |
ENST00000450849.7
|
ATCAY
|
ATCAY kinesin light chain interacting caytaxin |
| chr11_-_35419098 | 1.86 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr4_+_155903688 | 1.82 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr3_-_171026709 | 1.79 |
ENST00000314251.8
|
SLC2A2
|
solute carrier family 2 member 2 |
| chr3_-_149221811 | 1.77 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr2_-_49974083 | 1.77 |
ENST00000636345.1
|
NRXN1
|
neurexin 1 |
| chr9_+_72114595 | 1.74 |
ENST00000545168.5
|
GDA
|
guanine deaminase |
| chr17_-_39607876 | 1.71 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chr2_+_165239432 | 1.71 |
ENST00000636071.2
ENST00000636985.2 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr5_-_157345645 | 1.67 |
ENST00000312349.5
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr2_-_49974155 | 1.66 |
ENST00000635519.1
|
NRXN1
|
neurexin 1 |
| chr1_-_110607307 | 1.66 |
ENST00000639048.2
ENST00000675391.1 ENST00000639233.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr5_+_161850597 | 1.64 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr7_-_29969232 | 1.63 |
ENST00000409497.5
|
SCRN1
|
secernin 1 |
| chr9_+_74615582 | 1.62 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr2_-_49974182 | 1.62 |
ENST00000412315.5
ENST00000378262.7 |
NRXN1
|
neurexin 1 |
| chr4_-_20984011 | 1.53 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr5_+_127649018 | 1.53 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr16_+_72054477 | 1.46 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr13_-_83882456 | 1.45 |
ENST00000674365.1
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chr1_+_198639162 | 1.44 |
ENST00000418674.1
|
PTPRC
|
protein tyrosine phosphatase receptor type C |
| chr3_+_52777580 | 1.42 |
ENST00000273283.7
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr2_-_207166159 | 1.41 |
ENST00000426163.5
|
KLF7
|
Kruppel like factor 7 |
| chr2_+_165239388 | 1.38 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr10_+_66926028 | 1.35 |
ENST00000361320.5
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr18_-_28036585 | 1.32 |
ENST00000399380.7
|
CDH2
|
cadherin 2 |
| chr2_-_55419565 | 1.32 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr7_-_108328477 | 1.29 |
ENST00000442580.5
|
NRCAM
|
neuronal cell adhesion molecule |
| chr11_-_35418966 | 1.28 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr13_-_83882390 | 1.26 |
ENST00000377084.3
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chr4_+_157220691 | 1.25 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr1_-_115841116 | 1.24 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chr4_+_157220654 | 1.24 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr1_-_110607425 | 1.24 |
ENST00000633222.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr3_+_119782094 | 1.20 |
ENST00000393716.8
|
NR1I2
|
nuclear receptor subfamily 1 group I member 2 |
| chr3_-_186362223 | 1.17 |
ENST00000265022.8
|
DGKG
|
diacylglycerol kinase gamma |
| chr5_-_160852200 | 1.15 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr4_-_68670648 | 1.13 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr7_-_126533850 | 1.09 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr1_+_207089233 | 1.08 |
ENST00000243611.9
ENST00000367076.7 |
C4BPB
|
complement component 4 binding protein beta |
| chr2_-_55419276 | 1.05 |
ENST00000646796.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr12_+_20695323 | 1.01 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr1_+_196977550 | 1.00 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr7_-_108328635 | 1.00 |
ENST00000456431.5
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_-_110607970 | 0.99 |
ENST00000638532.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr2_+_214411048 | 0.98 |
ENST00000312504.10
ENST00000427124.1 |
VWC2L
|
von Willebrand factor C domain containing 2 like |
| chr3_+_122055355 | 0.96 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr7_-_29969927 | 0.96 |
ENST00000438497.5
|
SCRN1
|
secernin 1 |
| chr8_-_18684033 | 0.93 |
ENST00000614430.3
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr7_+_130266847 | 0.93 |
ENST00000222481.9
|
CPA2
|
carboxypeptidase A2 |
| chr8_+_53851786 | 0.93 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr1_+_207089283 | 0.93 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein beta |
| chr4_+_186227501 | 0.92 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr2_-_60546162 | 0.92 |
ENST00000489183.1
ENST00000647469.1 ENST00000642180.1 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr12_+_20695442 | 0.91 |
ENST00000540354.5
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr8_+_103819212 | 0.91 |
ENST00000515551.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_+_20695469 | 0.89 |
ENST00000545604.5
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr8_-_18684093 | 0.89 |
ENST00000428502.6
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_+_206557157 | 0.89 |
ENST00000577571.5
|
RASSF5
|
Ras association domain family member 5 |
| chr1_+_207089195 | 0.88 |
ENST00000452902.6
|
C4BPB
|
complement component 4 binding protein beta |
| chr5_-_157345559 | 0.88 |
ENST00000520782.1
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr1_+_92168915 | 0.87 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr8_+_18391276 | 0.87 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr4_-_101346842 | 0.87 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr1_-_110607816 | 0.86 |
ENST00000485317.6
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr7_+_129144691 | 0.85 |
ENST00000486685.3
|
TSPAN33
|
tetraspanin 33 |
| chr2_-_49973939 | 0.85 |
ENST00000630656.1
|
NRXN1
|
neurexin 1 |
| chr4_-_101347471 | 0.82 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101347327 | 0.82 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr12_-_56752311 | 0.81 |
ENST00000338193.11
ENST00000550770.1 |
PRIM1
|
DNA primase subunit 1 |
| chr1_-_243163310 | 0.80 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr1_+_116754422 | 0.80 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr4_-_101347492 | 0.78 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr2_-_162838728 | 0.77 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr7_+_120273129 | 0.76 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr7_+_142791635 | 0.75 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chrX_-_120559889 | 0.73 |
ENST00000371323.3
|
CUL4B
|
cullin 4B |
| chr4_-_101347551 | 0.71 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr15_-_76059755 | 0.68 |
ENST00000563910.5
|
NRG4
|
neuregulin 4 |
| chr6_+_13013513 | 0.67 |
ENST00000675203.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr3_-_120093666 | 0.67 |
ENST00000316626.6
ENST00000650344.2 ENST00000678439.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chr17_-_4263847 | 0.67 |
ENST00000570535.5
ENST00000574367.5 ENST00000341657.9 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr12_-_56752366 | 0.66 |
ENST00000672280.1
|
PRIM1
|
DNA primase subunit 1 |
| chr19_-_48513161 | 0.64 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chrX_+_10156960 | 0.63 |
ENST00000380833.9
|
CLCN4
|
chloride voltage-gated channel 4 |
| chr12_+_100200779 | 0.60 |
ENST00000188312.7
ENST00000546902.5 ENST00000552376.5 ENST00000551617.1 |
ACTR6
|
actin related protein 6 |
| chr19_+_35449584 | 0.60 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr21_-_14546297 | 0.59 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr11_-_60855943 | 0.57 |
ENST00000332539.5
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr7_-_28180735 | 0.57 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr16_+_7303245 | 0.57 |
ENST00000674626.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr7_+_142450941 | 0.57 |
ENST00000390368.2
|
TRBV6-5
|
T cell receptor beta variable 6-5 |
| chr2_+_167868948 | 0.57 |
ENST00000392690.3
|
B3GALT1
|
beta-1,3-galactosyltransferase 1 |
| chr21_-_14546351 | 0.56 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chrX_+_47233772 | 0.53 |
ENST00000377078.2
|
USP11
|
ubiquitin specific peptidase 11 |
| chr16_-_28181034 | 0.53 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr2_+_33436304 | 0.53 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr15_+_47759723 | 0.52 |
ENST00000559196.1
|
SEMA6D
|
semaphorin 6D |
| chr9_-_28670285 | 0.52 |
ENST00000379992.6
ENST00000308675.5 ENST00000613945.3 |
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_+_166989089 | 0.52 |
ENST00000367870.6
|
MAEL
|
maelstrom spermatogenic transposon silencer |
| chr12_+_8992029 | 0.51 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr10_-_60572599 | 0.51 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr8_+_32647080 | 0.50 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr12_+_80099535 | 0.49 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr2_-_207165923 | 0.49 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr4_+_56216101 | 0.48 |
ENST00000504228.6
|
CRACD
|
capping protein inhibiting regulator of actin dynamics |
| chr7_-_78771265 | 0.47 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_+_186266183 | 0.47 |
ENST00000403665.7
ENST00000492972.6 ENST00000264692.8 |
F11
|
coagulation factor XI |
| chr3_-_24165548 | 0.46 |
ENST00000280696.9
|
THRB
|
thyroid hormone receptor beta |
| chr14_+_39267377 | 0.45 |
ENST00000556148.5
ENST00000348007.7 |
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr3_+_159839847 | 0.45 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr17_-_2401038 | 0.45 |
ENST00000174618.5
ENST00000575394.1 |
MNT
|
MAX network transcriptional repressor |
| chr1_+_113930728 | 0.44 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr6_-_152563271 | 0.44 |
ENST00000535896.7
ENST00000672122.1 |
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr17_+_30378903 | 0.43 |
ENST00000225719.9
|
CPD
|
carboxypeptidase D |
| chr7_-_151633182 | 0.43 |
ENST00000476632.2
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr3_-_194398354 | 0.43 |
ENST00000401815.1
|
GP5
|
glycoprotein V platelet |
| chr17_+_7558296 | 0.42 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr14_+_21797272 | 0.41 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr3_+_134795248 | 0.40 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chr12_+_59596137 | 0.39 |
ENST00000549465.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr8_-_144605699 | 0.39 |
ENST00000377307.6
ENST00000276826.5 |
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr15_+_66386902 | 0.38 |
ENST00000307102.10
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr8_+_31639755 | 0.38 |
ENST00000520407.5
|
NRG1
|
neuregulin 1 |
| chr12_-_121580201 | 0.38 |
ENST00000539394.5
|
KDM2B
|
lysine demethylase 2B |
| chr8_+_76681208 | 0.36 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr9_-_74952904 | 0.36 |
ENST00000376854.6
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr5_-_62403506 | 0.36 |
ENST00000680062.1
|
DIMT1
|
DIMT1 rRNA methyltransferase and ribosome maturation factor |
| chr12_-_121579638 | 0.36 |
ENST00000446152.6
|
KDM2B
|
lysine demethylase 2B |
| chr6_-_152563349 | 0.35 |
ENST00000673281.1
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr12_+_32679200 | 0.35 |
ENST00000452533.6
ENST00000414834.6 |
DNM1L
|
dynamin 1 like |
| chr14_+_69398683 | 0.34 |
ENST00000556605.5
ENST00000031146.8 ENST00000336643.10 |
SLC39A9
|
solute carrier family 39 member 9 |
| chr10_-_96271508 | 0.34 |
ENST00000427367.6
ENST00000413476.6 ENST00000371176.6 |
BLNK
|
B cell linker |
| chr11_+_64917561 | 0.34 |
ENST00000526559.5
|
PPP2R5B
|
protein phosphatase 2 regulatory subunit B'beta |
| chrX_-_64976435 | 0.33 |
ENST00000374839.8
|
ZC4H2
|
zinc finger C4H2-type containing |
| chr1_-_151459471 | 0.32 |
ENST00000271715.7
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr19_+_7534004 | 0.32 |
ENST00000221249.10
ENST00000601668.5 ENST00000601001.5 |
PNPLA6
|
patatin like phospholipase domain containing 6 |
| chr17_+_17782108 | 0.31 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr17_-_72992933 | 0.31 |
ENST00000582769.5
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr6_+_147204525 | 0.31 |
ENST00000321680.10
ENST00000367480.7 |
STXBP5
|
syntaxin binding protein 5 |
| chr1_+_154405193 | 0.31 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr4_+_70721953 | 0.30 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr5_-_83673544 | 0.30 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr14_-_54441325 | 0.30 |
ENST00000556113.1
ENST00000553660.5 ENST00000216416.9 ENST00000395573.8 ENST00000557690.5 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr12_+_41437680 | 0.29 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr3_-_155676363 | 0.29 |
ENST00000494598.5
|
PLCH1
|
phospholipase C eta 1 |
| chr7_+_99828010 | 0.29 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr9_+_92963961 | 0.29 |
ENST00000416701.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr2_+_6877768 | 0.28 |
ENST00000382040.4
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr10_-_125816596 | 0.28 |
ENST00000368786.5
|
UROS
|
uroporphyrinogen III synthase |
| chr1_-_151459198 | 0.28 |
ENST00000531094.5
|
POGZ
|
pogo transposable element derived with ZNF domain |
| chr3_-_131026558 | 0.27 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 |
| chr6_+_113857333 | 0.27 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr22_+_30080460 | 0.26 |
ENST00000336726.11
|
HORMAD2
|
HORMA domain containing 2 |
| chr10_-_119536533 | 0.26 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr16_-_30610342 | 0.25 |
ENST00000287461.8
|
ZNF689
|
zinc finger protein 689 |
| chr2_-_213152427 | 0.25 |
ENST00000452786.2
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr16_+_25216943 | 0.25 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chr3_-_94028582 | 0.24 |
ENST00000315099.3
|
STX19
|
syntaxin 19 |
| chr12_+_32679269 | 0.23 |
ENST00000358214.9
ENST00000553257.6 ENST00000549701.6 ENST00000266481.10 ENST00000551476.5 ENST00000550154.5 ENST00000547312.5 ENST00000381000.8 ENST00000548750.5 |
DNM1L
|
dynamin 1 like |
| chr12_+_48472559 | 0.23 |
ENST00000266594.4
|
ANP32D
|
acidic nuclear phosphoprotein 32 family member D |
| chr18_+_63542365 | 0.22 |
ENST00000269491.6
ENST00000382768.2 |
SERPINB12
|
serpin family B member 12 |
| chr3_+_109136707 | 0.22 |
ENST00000622536.6
|
C3orf85
|
chromosome 3 open reading frame 85 |
| chr6_-_151452018 | 0.21 |
ENST00000491268.2
|
RMND1
|
required for meiotic nuclear division 1 homolog |
| chr1_+_149475933 | 0.21 |
ENST00000612881.4
|
NBPF19
|
NBPF member 19 |
| chr1_+_154405573 | 0.20 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr6_-_75284820 | 0.20 |
ENST00000475111.6
|
TMEM30A
|
transmembrane protein 30A |
| chr4_+_70050431 | 0.20 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.9 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 1.7 | 5.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.8 | 5.0 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.8 | 4.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.6 | 1.8 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.6 | 1.7 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.6 | 3.4 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.5 | 1.5 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.5 | 1.9 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.5 | 4.7 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.4 | 1.7 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.4 | 2.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 2.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 5.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.4 | 2.7 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.4 | 1.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.3 | 1.4 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.3 | 5.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.0 | GO:0032681 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.3 | 2.9 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 1.9 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.2 | 15.0 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.2 | 1.9 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 1.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 4.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.6 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.2 | 2.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 3.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.2 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 2.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 2.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.2 | 3.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 0.9 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.9 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 2.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.7 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.4 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.1 | 1.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 2.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 6.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.7 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.8 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.1 | 1.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.3 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.5 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 1.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.8 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.4 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 1.2 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 2.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 1.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.8 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 9.7 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 3.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.9 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 3.6 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.5 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 1.2 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 2.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.9 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 2.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.3 | 4.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 10.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 5.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.3 | 4.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 4.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 14.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.5 | GO:0030849 | autosome(GO:0030849) |
| 0.1 | 3.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 2.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 5.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 2.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 7.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 8.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 12.4 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 2.9 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 15.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.9 | 8.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 15.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.6 | 1.7 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.4 | 5.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.8 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 2.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 4.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.3 | 1.5 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 2.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 0.9 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.3 | 2.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 8.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 5.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 2.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 0.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 5.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 2.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.5 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.4 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.1 | 2.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 1.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 2.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 8.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 1.1 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 9.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 9.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 3.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 17.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 3.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 6.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 4.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 5.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |