Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for OSR1_OSR2
Z-value: 1.27
Transcription factors associated with OSR1_OSR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
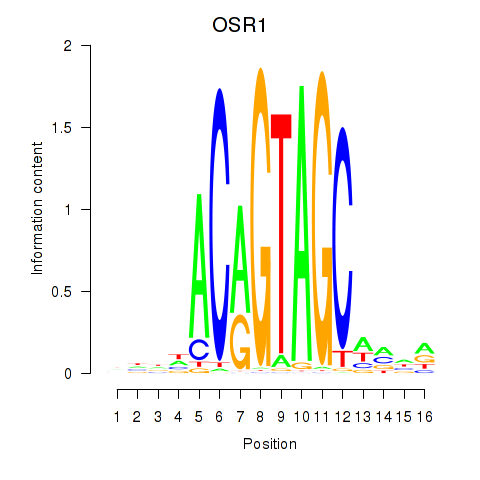
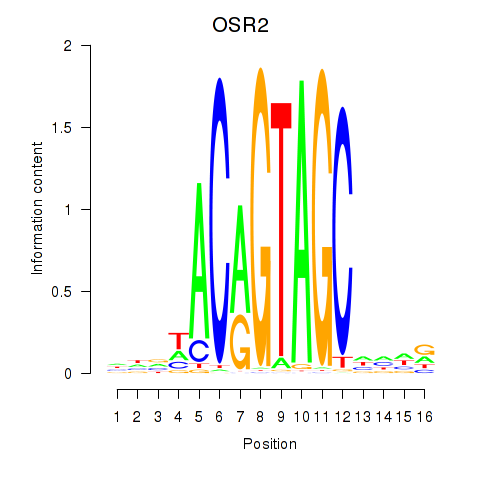
OSR1
|
ENSG00000143867.7 | OSR1 |
|
OSR2
|
ENSG00000164920.9 | OSR2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OSR1 | hg38_v1_chr2_-_19358612_19358631 | 0.03 | 8.8e-01 | Click! |
| OSR2 | hg38_v1_chr8_+_98944403_98944458 | -0.02 | 9.1e-01 | Click! |
Activity profile of OSR1_OSR2 motif
Sorted Z-values of OSR1_OSR2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OSR1_OSR2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_69096494 | 8.98 |
ENST00000508661.5
ENST00000622664.1 |
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr4_+_69096467 | 8.51 |
ENST00000305231.12
|
UGT2B7
|
UDP glucuronosyltransferase family 2 member B7 |
| chr4_-_69495897 | 2.55 |
ENST00000305107.7
ENST00000639621.1 |
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr7_+_134527560 | 2.50 |
ENST00000359579.5
|
AKR1B10
|
aldo-keto reductase family 1 member B10 |
| chr4_-_69495861 | 2.28 |
ENST00000512583.5
|
UGT2B4
|
UDP glucuronosyltransferase family 2 member B4 |
| chr16_+_89630263 | 2.26 |
ENST00000261615.5
|
DPEP1
|
dipeptidase 1 |
| chr4_+_168092530 | 2.23 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr2_+_233671879 | 2.20 |
ENST00000354728.5
|
UGT1A9
|
UDP glucuronosyltransferase family 1 member A9 |
| chr2_-_87825952 | 2.10 |
ENST00000398146.4
|
RGPD2
|
RANBP2 like and GRIP domain containing 2 |
| chr16_-_56668034 | 2.09 |
ENST00000569500.5
ENST00000379811.4 ENST00000444837.6 |
MT1G
|
metallothionein 1G |
| chr19_-_45322867 | 2.01 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr1_-_151369704 | 1.96 |
ENST00000424475.5
|
SELENBP1
|
selenium binding protein 1 |
| chr14_+_20684177 | 1.92 |
ENST00000336811.10
|
ANG
|
angiogenin |
| chr3_+_149474688 | 1.75 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr8_+_65707041 | 1.67 |
ENST00000521247.6
ENST00000527155.5 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr7_-_95596507 | 1.65 |
ENST00000005178.6
|
PDK4
|
pyruvate dehydrogenase kinase 4 |
| chr4_-_69214743 | 1.54 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr2_+_234050679 | 1.37 |
ENST00000373368.5
ENST00000168148.8 |
SPP2
|
secreted phosphoprotein 2 |
| chr1_-_206970457 | 1.27 |
ENST00000324852.9
ENST00000450945.3 ENST00000400962.8 |
FCAMR
|
Fc fragment of IgA and IgM receptor |
| chr9_-_34710069 | 1.25 |
ENST00000378792.1
ENST00000259607.7 |
CCL21
|
C-C motif chemokine ligand 21 |
| chr17_-_48613468 | 1.14 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr11_-_72794032 | 1.14 |
ENST00000334805.11
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chrX_-_42778155 | 1.04 |
ENST00000378131.4
|
PPP1R2C
|
PPP1R2C family member C |
| chr12_+_108880085 | 1.03 |
ENST00000228476.8
ENST00000547768.5 |
DAO
|
D-amino acid oxidase |
| chr14_+_20684547 | 1.01 |
ENST00000555835.3
ENST00000397995.2 ENST00000553909.1 |
RNASE4
ENSG00000259171.1
|
ribonuclease A family member 4 novel protein, ANG-RNASE4 readthrough |
| chr2_+_234050732 | 0.96 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2 |
| chr1_-_58577244 | 0.95 |
ENST00000371225.4
|
TACSTD2
|
tumor associated calcium signal transducer 2 |
| chr4_-_185775271 | 0.94 |
ENST00000430503.5
ENST00000319454.10 ENST00000450341.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_42887392 | 0.90 |
ENST00000514218.5
|
SELENOP
|
selenoprotein P |
| chr22_-_23632317 | 0.85 |
ENST00000317749.9
|
DRICH1
|
aspartate rich 1 |
| chr18_+_11981015 | 0.83 |
ENST00000589238.5
|
IMPA2
|
inositol monophosphatase 2 |
| chr3_+_50205254 | 0.81 |
ENST00000614032.5
ENST00000445096.5 |
SLC38A3
|
solute carrier family 38 member 3 |
| chr15_+_58431985 | 0.78 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr4_+_39459039 | 0.78 |
ENST00000509519.5
ENST00000640888.2 ENST00000424936.6 ENST00000381846.2 ENST00000513731.6 |
LIAS
|
lipoic acid synthetase |
| chr22_-_26590082 | 0.77 |
ENST00000442495.5
ENST00000440953.5 ENST00000450022.1 ENST00000338754.9 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr9_+_131009141 | 0.73 |
ENST00000361069.9
|
LAMC3
|
laminin subunit gamma 3 |
| chr1_+_196888014 | 0.73 |
ENST00000367416.6
ENST00000608469.6 ENST00000251424.8 ENST00000367418.2 |
CFHR4
|
complement factor H related 4 |
| chr7_+_130207847 | 0.72 |
ENST00000297819.4
|
SSMEM1
|
serine rich single-pass membrane protein 1 |
| chr2_+_37231761 | 0.72 |
ENST00000416653.5
|
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr15_+_36702009 | 0.71 |
ENST00000562489.1
|
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr18_-_50195138 | 0.70 |
ENST00000285039.12
|
MYO5B
|
myosin VB |
| chr2_+_48617841 | 0.70 |
ENST00000403751.8
|
GTF2A1L
|
general transcription factor IIA subunit 1 like |
| chr8_-_80029904 | 0.67 |
ENST00000521434.5
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr3_-_50322759 | 0.67 |
ENST00000442581.1
ENST00000447092.5 |
HYAL2
|
hyaluronidase 2 |
| chr2_+_48617798 | 0.66 |
ENST00000448460.5
ENST00000437125.5 ENST00000430487.6 |
GTF2A1L
|
general transcription factor IIA subunit 1 like |
| chr1_-_119896489 | 0.64 |
ENST00000369400.2
|
ADAM30
|
ADAM metallopeptidase domain 30 |
| chr16_-_66934144 | 0.63 |
ENST00000568572.5
|
CIAO2B
|
cytosolic iron-sulfur assembly component 2B |
| chr2_+_37231633 | 0.62 |
ENST00000002125.9
ENST00000336237.10 ENST00000431821.5 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr2_+_37231798 | 0.62 |
ENST00000439218.5
ENST00000432075.1 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr3_+_194136138 | 0.62 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr10_-_77637902 | 0.61 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr6_+_10585748 | 0.60 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr10_-_77637721 | 0.58 |
ENST00000638848.1
ENST00000639406.1 ENST00000618048.2 ENST00000639120.1 ENST00000640834.1 ENST00000639601.1 ENST00000638514.1 ENST00000457953.6 ENST00000639090.1 ENST00000639489.1 ENST00000372440.6 ENST00000404771.8 ENST00000638203.1 ENST00000638306.1 ENST00000638351.1 ENST00000638606.1 ENST00000639591.1 ENST00000640182.1 ENST00000640605.1 ENST00000640141.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr15_+_73443149 | 0.56 |
ENST00000560581.1
ENST00000331090.11 |
REC114
|
REC114 meiotic recombination protein |
| chr17_-_81937320 | 0.54 |
ENST00000577624.5
ENST00000403172.8 ENST00000619204.4 ENST00000629768.2 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr18_-_22417910 | 0.54 |
ENST00000391403.4
|
CTAGE1
|
cutaneous T cell lymphoma-associated antigen 1 |
| chr3_+_50205240 | 0.53 |
ENST00000610458.4
|
SLC38A3
|
solute carrier family 38 member 3 |
| chr4_-_185775890 | 0.53 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_-_44065541 | 0.52 |
ENST00000297283.4
|
PGAM2
|
phosphoglycerate mutase 2 |
| chr4_+_39459069 | 0.52 |
ENST00000261434.8
ENST00000340169.7 ENST00000638451.1 ENST00000640349.1 |
LIAS
|
lipoic acid synthetase |
| chr10_-_77637789 | 0.51 |
ENST00000481070.1
ENST00000640969.1 ENST00000286628.14 ENST00000638991.1 ENST00000639913.1 ENST00000480683.2 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr3_-_46812558 | 0.49 |
ENST00000641183.1
ENST00000460241.2 |
ENSG00000284672.1
ENSG00000206549.13
|
novel transcript novel protein identical to PRSS50 |
| chr3_-_50322733 | 0.49 |
ENST00000428028.1
ENST00000357750.9 |
HYAL2
|
hyaluronidase 2 |
| chr4_+_150582119 | 0.48 |
ENST00000317605.6
|
MAB21L2
|
mab-21 like 2 |
| chr12_-_14814160 | 0.47 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr14_+_58395912 | 0.47 |
ENST00000360945.7
|
TOMM20L
|
translocase of outer mitochondrial membrane 20 like |
| chrX_-_139965510 | 0.47 |
ENST00000370540.2
|
CXorf66
|
chromosome X open reading frame 66 |
| chr2_+_219491867 | 0.47 |
ENST00000412982.5
|
ENSG00000286143.1
|
novel protein |
| chr10_-_77637558 | 0.47 |
ENST00000372421.10
ENST00000639370.1 ENST00000640773.1 ENST00000638895.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr14_+_64986846 | 0.46 |
ENST00000246166.3
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr8_-_80029826 | 0.46 |
ENST00000519386.5
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr12_-_57772087 | 0.46 |
ENST00000324871.12
ENST00000257848.7 |
METTL1
|
methyltransferase like 1 |
| chr7_+_73692596 | 0.46 |
ENST00000453316.1
|
BUD23
|
BUD23 rRNA methyltransferase and ribosome maturation factor |
| chr4_-_185775411 | 0.45 |
ENST00000445115.5
ENST00000451701.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_-_81201886 | 0.44 |
ENST00000451455.1
ENST00000358130.7 ENST00000436386.5 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr4_-_185775376 | 0.44 |
ENST00000456596.5
ENST00000414724.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_141719162 | 0.43 |
ENST00000493845.1
|
WEE2
|
WEE2 oocyte meiosis inhibiting kinase |
| chr7_+_123925236 | 0.42 |
ENST00000413927.5
ENST00000682466.1 ENST00000460182.5 ENST00000340011.9 |
SPAM1
|
sperm adhesion molecule 1 |
| chr4_-_40514543 | 0.42 |
ENST00000513473.5
|
RBM47
|
RNA binding motif protein 47 |
| chr1_-_21622509 | 0.42 |
ENST00000374761.6
|
RAP1GAP
|
RAP1 GTPase activating protein |
| chr10_-_77637633 | 0.42 |
ENST00000638223.1
ENST00000639544.1 ENST00000640807.1 ENST00000434208.6 ENST00000626620.3 ENST00000638575.1 ENST00000638759.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_46798998 | 0.42 |
ENST00000640628.1
ENST00000271153.8 ENST00000371923.9 ENST00000371919.8 ENST00000614163.4 |
CYP4B1
|
cytochrome P450 family 4 subfamily B member 1 |
| chr12_-_21775045 | 0.40 |
ENST00000667884.1
|
KCNJ8
|
potassium inwardly rectifying channel subfamily J member 8 |
| chr15_-_50765656 | 0.40 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr19_+_35712928 | 0.39 |
ENST00000262630.7
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr21_+_38296909 | 0.39 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr17_-_81937221 | 0.38 |
ENST00000402252.6
ENST00000583564.5 ENST00000585244.1 ENST00000337943.9 ENST00000579698.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr17_-_81937277 | 0.38 |
ENST00000405481.8
ENST00000329875.13 ENST00000585215.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr9_+_37650947 | 0.37 |
ENST00000377765.8
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr20_+_38581186 | 0.37 |
ENST00000373348.4
ENST00000537425.2 ENST00000416116.2 |
ADIG
|
adipogenin |
| chr22_+_37024137 | 0.36 |
ENST00000628507.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr20_-_35411963 | 0.35 |
ENST00000349714.9
ENST00000438533.5 ENST00000359226.6 ENST00000374384.6 ENST00000374385.10 ENST00000424405.5 ENST00000397554.5 ENST00000374380.6 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr4_-_993430 | 0.35 |
ENST00000361661.6
ENST00000622731.4 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr3_-_179604648 | 0.34 |
ENST00000392659.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr2_-_96505345 | 0.33 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr10_-_77637444 | 0.33 |
ENST00000639205.1
ENST00000639498.1 ENST00000372408.7 ENST00000372403.9 ENST00000640626.1 ENST00000404857.6 ENST00000638252.1 ENST00000640029.1 ENST00000640934.1 ENST00000639823.1 ENST00000372437.6 ENST00000639344.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr19_-_56197793 | 0.32 |
ENST00000589938.5
ENST00000587032.2 ENST00000586855.6 |
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chrX_-_15315615 | 0.32 |
ENST00000380470.7
ENST00000480796.6 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr5_-_178187364 | 0.32 |
ENST00000463439.3
|
GMCL2
|
germ cell-less 2, spermatogenesis associated |
| chr11_-_8682619 | 0.31 |
ENST00000646038.2
|
TRIM66
|
tripartite motif containing 66 |
| chr4_-_185775484 | 0.31 |
ENST00000444771.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_179604628 | 0.30 |
ENST00000476781.6
ENST00000259038.6 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr4_-_109729956 | 0.30 |
ENST00000502283.1
|
PLA2G12A
|
phospholipase A2 group XIIA |
| chr4_-_993376 | 0.28 |
ENST00000398520.6
ENST00000398516.3 |
SLC26A1
|
solute carrier family 26 member 1 |
| chr6_-_136466858 | 0.27 |
ENST00000544465.5
|
MAP7
|
microtubule associated protein 7 |
| chr8_+_69466837 | 0.27 |
ENST00000529134.5
|
SULF1
|
sulfatase 1 |
| chr7_-_6348758 | 0.27 |
ENST00000578372.1
|
ENSG00000286075.2
|
novel protein |
| chrX_+_70062457 | 0.27 |
ENST00000338352.3
|
OTUD6A
|
OTU deubiquitinase 6A |
| chr12_-_86256267 | 0.26 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr10_-_43409160 | 0.26 |
ENST00000337970.7
ENST00000682386.1 |
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr4_-_185775432 | 0.26 |
ENST00000457247.5
ENST00000435480.5 ENST00000425679.5 ENST00000457934.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_-_65842051 | 0.25 |
ENST00000401827.8
|
PDE7A
|
phosphodiesterase 7A |
| chrY_+_3579067 | 0.24 |
ENST00000321217.5
ENST00000559055.2 |
TGIF2LY
|
TGFB induced factor homeobox 2 like Y-linked |
| chr17_+_42562120 | 0.24 |
ENST00000585811.1
ENST00000585909.1 ENST00000586771.1 ENST00000421097.6 ENST00000591779.5 ENST00000393818.3 ENST00000587858.5 ENST00000587214.1 ENST00000587157.1 ENST00000590958.5 |
COASY
|
Coenzyme A synthase |
| chr3_+_50617390 | 0.23 |
ENST00000457064.1
|
MAPKAPK3
|
MAPK activated protein kinase 3 |
| chr2_-_189583392 | 0.23 |
ENST00000440626.1
|
SLC40A1
|
solute carrier family 40 member 1 |
| chr19_-_5680191 | 0.23 |
ENST00000590389.5
|
MICOS13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chr15_+_41231219 | 0.23 |
ENST00000334660.10
ENST00000560397.5 |
CHP1
|
calcineurin like EF-hand protein 1 |
| chr18_+_65750628 | 0.23 |
ENST00000536984.6
|
CDH7
|
cadherin 7 |
| chr9_-_33025088 | 0.22 |
ENST00000436040.7
|
APTX
|
aprataxin |
| chr19_-_51531790 | 0.22 |
ENST00000359982.8
ENST00000436458.5 ENST00000391797.3 ENST00000343300.8 |
SIGLEC6
|
sialic acid binding Ig like lectin 6 |
| chr20_+_35954564 | 0.22 |
ENST00000622112.4
ENST00000614708.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr9_+_36572854 | 0.21 |
ENST00000543751.5
ENST00000536860.5 ENST00000541717.4 ENST00000536329.5 ENST00000536987.5 ENST00000298048.7 ENST00000495529.5 ENST00000545008.5 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr12_-_103957122 | 0.21 |
ENST00000552940.1
ENST00000547975.5 ENST00000549478.1 ENST00000546540.1 ENST00000378090.9 ENST00000546819.1 ENST00000547945.5 |
C12orf73
|
chromosome 12 open reading frame 73 |
| chrX_-_13319952 | 0.20 |
ENST00000622204.1
ENST00000380622.5 |
ATXN3L
|
ataxin 3 like |
| chr17_-_55511434 | 0.20 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36 |
| chr12_-_86256299 | 0.20 |
ENST00000552808.6
ENST00000547225.5 |
MGAT4C
|
MGAT4 family member C |
| chr12_-_86256376 | 0.20 |
ENST00000552435.6
|
MGAT4C
|
MGAT4 family member C |
| chr19_-_12957198 | 0.19 |
ENST00000316939.3
|
GADD45GIP1
|
GADD45G interacting protein 1 |
| chr2_-_182522556 | 0.19 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A |
| chr17_+_41232447 | 0.19 |
ENST00000411528.4
|
KRTAP9-3
|
keratin associated protein 9-3 |
| chr19_-_5680220 | 0.19 |
ENST00000587950.5
|
MICOS13
|
mitochondrial contact site and cristae organizing system subunit 13 |
| chrX_-_15384402 | 0.18 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D |
| chr19_+_55080363 | 0.18 |
ENST00000588359.5
ENST00000245618.5 |
EPS8L1
|
EPS8 like 1 |
| chr12_-_101210232 | 0.18 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8 |
| chr10_+_52128343 | 0.17 |
ENST00000672084.1
|
PRKG1
|
protein kinase cGMP-dependent 1 |
| chr16_-_8868343 | 0.17 |
ENST00000562843.5
ENST00000561530.5 ENST00000396593.6 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr6_+_108295037 | 0.16 |
ENST00000368977.9
ENST00000421954.5 |
AFG1L
|
AFG1 like ATPase |
| chr12_-_16277685 | 0.16 |
ENST00000344941.3
|
SLC15A5
|
solute carrier family 15 member 5 |
| chr11_-_114559847 | 0.16 |
ENST00000251921.6
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1 |
| chr10_-_43408787 | 0.16 |
ENST00000356053.7
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_-_341417 | 0.16 |
ENST00000575634.5
|
RPH3AL
|
rabphilin 3A like (without C2 domains) |
| chr1_-_34985288 | 0.16 |
ENST00000417456.1
ENST00000373337.3 |
ENSG00000284773.1
TMEM35B
|
novel protein transmembrane protein 35B |
| chr1_-_196608359 | 0.16 |
ENST00000609185.5
ENST00000451324.6 ENST00000367433.9 ENST00000294725.14 |
KCNT2
|
potassium sodium-activated channel subfamily T member 2 |
| chrX_+_9912548 | 0.15 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr21_-_33542841 | 0.15 |
ENST00000381831.7
ENST00000381839.7 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr10_-_72354895 | 0.15 |
ENST00000444643.8
ENST00000338820.7 ENST00000394903.6 |
DNAJB12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr2_+_188291661 | 0.15 |
ENST00000409843.5
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr16_+_67109977 | 0.15 |
ENST00000566026.1
|
C16orf70
|
chromosome 16 open reading frame 70 |
| chr17_-_81936775 | 0.15 |
ENST00000584848.5
ENST00000577756.5 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr12_+_57772648 | 0.15 |
ENST00000333012.5
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr4_+_145482761 | 0.15 |
ENST00000507367.1
ENST00000394092.6 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr19_+_30228530 | 0.15 |
ENST00000591488.1
|
ZNF536
|
zinc finger protein 536 |
| chr7_-_127618137 | 0.14 |
ENST00000639438.3
|
PAX4
|
paired box 4 |
| chr13_+_52652828 | 0.14 |
ENST00000310528.9
ENST00000343788.10 |
SUGT1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr19_+_23914873 | 0.14 |
ENST00000525354.6
ENST00000334589.9 ENST00000531821.6 ENST00000322487.11 ENST00000594466.6 |
ZNF726
|
zinc finger protein 726 |
| chr19_-_51531717 | 0.14 |
ENST00000346477.7
|
SIGLEC6
|
sialic acid binding Ig like lectin 6 |
| chr3_-_45915596 | 0.14 |
ENST00000472635.5
ENST00000492333.5 |
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr1_+_13061158 | 0.14 |
ENST00000681473.1
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr19_+_44955365 | 0.14 |
ENST00000337392.10
ENST00000591304.1 |
CLPTM1
|
CLPTM1 regulator of GABA type A receptor forward trafficking |
| chr13_-_71867192 | 0.14 |
ENST00000611519.4
ENST00000620444.4 ENST00000613252.5 |
DACH1
|
dachshund family transcription factor 1 |
| chr5_-_140673568 | 0.13 |
ENST00000542735.2
|
DND1
|
DND microRNA-mediated repression inhibitor 1 |
| chr1_+_37807762 | 0.13 |
ENST00000373042.5
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr12_+_28452493 | 0.13 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr6_+_27138588 | 0.12 |
ENST00000615353.1
|
H4C9
|
H4 clustered histone 9 |
| chr3_-_172523423 | 0.12 |
ENST00000241261.7
|
TNFSF10
|
TNF superfamily member 10 |
| chr1_+_156591741 | 0.12 |
ENST00000368234.7
ENST00000680087.1 ENST00000681734.1 ENST00000679369.1 ENST00000680269.1 ENST00000680661.1 ENST00000681054.1 ENST00000680004.1 ENST00000679702.1 ENST00000368235.8 ENST00000368233.3 |
NAXE
|
NAD(P)HX epimerase |
| chr1_-_13165631 | 0.12 |
ENST00000323770.8
|
HNRNPCL4
|
heterogeneous nuclear ribonucleoprotein C like 4 |
| chr11_-_8682680 | 0.12 |
ENST00000644261.1
|
TRIM66
|
tripartite motif containing 66 |
| chr17_-_74872961 | 0.11 |
ENST00000581530.5
ENST00000420580.6 ENST00000413947.6 ENST00000581219.1 ENST00000582944.5 ENST00000293195.10 ENST00000583917.5 ENST00000442102.6 |
FDXR
|
ferredoxin reductase |
| chr5_+_110738134 | 0.11 |
ENST00000513807.5
|
SLC25A46
|
solute carrier family 25 member 46 |
| chrX_-_154832671 | 0.11 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chr3_+_130929998 | 0.11 |
ENST00000508297.2
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr20_+_10435283 | 0.11 |
ENST00000334534.10
|
SLX4IP
|
SLX4 interacting protein |
| chr11_-_114559866 | 0.11 |
ENST00000534921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family member 1 |
| chr4_-_109729987 | 0.10 |
ENST00000243501.10
|
PLA2G12A
|
phospholipase A2 group XIIA |
| chr19_+_23914904 | 0.10 |
ENST00000575986.1
|
ZNF726
|
zinc finger protein 726 |
| chr3_-_172523460 | 0.10 |
ENST00000420541.6
|
TNFSF10
|
TNF superfamily member 10 |
| chr1_+_75796867 | 0.10 |
ENST00000263187.4
|
MSH4
|
mutS homolog 4 |
| chr10_-_28334439 | 0.10 |
ENST00000441595.2
|
MPP7
|
membrane palmitoylated protein 7 |
| chr14_+_21653835 | 0.10 |
ENST00000641524.1
|
OR4E2
|
olfactory receptor family 4 subfamily E member 2 |
| chr19_-_17075038 | 0.09 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin like complex subunit 8 |
| chr8_+_37736612 | 0.09 |
ENST00000518526.5
ENST00000523887.5 ENST00000648919.1 ENST00000519638.3 |
ERLIN2
|
ER lipid raft associated 2 |
| chr6_+_151690492 | 0.09 |
ENST00000404742.5
ENST00000440973.5 |
ESR1
|
estrogen receptor 1 |
| chr12_+_57772587 | 0.09 |
ENST00000300209.13
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr8_+_93698561 | 0.09 |
ENST00000523475.5
|
CIBAR1
|
CBY1 interacting BAR domain containing 1 |
| chrX_-_108736556 | 0.09 |
ENST00000372129.4
|
IRS4
|
insulin receptor substrate 4 |
| chr1_+_26177482 | 0.08 |
ENST00000361530.11
ENST00000374253.9 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr2_-_182522703 | 0.08 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr15_-_66356672 | 0.08 |
ENST00000261881.9
|
TIPIN
|
TIMELESS interacting protein |
| chr10_+_47348351 | 0.08 |
ENST00000584701.2
|
RBP3
|
retinol binding protein 3 |
| chr7_+_16661182 | 0.07 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr10_-_109923428 | 0.07 |
ENST00000403138.6
ENST00000369683.5 ENST00000502935.6 ENST00000322238.12 |
XPNPEP1
|
X-prolyl aminopeptidase 1 |
| chr11_+_6845683 | 0.07 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr19_+_44954577 | 0.07 |
ENST00000546079.5
ENST00000541297.6 |
CLPTM1
|
CLPTM1 regulator of GABA type A receptor forward trafficking |
| chrX_-_108736327 | 0.07 |
ENST00000564206.2
|
IRS4
|
insulin receptor substrate 4 |
| chr16_-_48385397 | 0.07 |
ENST00000394725.3
|
SIAH1
|
siah E3 ubiquitin protein ligase 1 |
| chr1_+_207645114 | 0.06 |
ENST00000508064.7
|
CR1L
|
complement C3b/C4b receptor 1 like |
| chr5_-_36151853 | 0.06 |
ENST00000296603.5
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr1_+_9943428 | 0.06 |
ENST00000403197.5
ENST00000377205.6 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.9 | 19.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.8 | 2.5 | GO:0016487 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.8 | 2.3 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.4 | 1.3 | GO:2000485 | asparagine transport(GO:0006867) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) regulation of glutamine transport(GO:2000485) |
| 0.4 | 1.2 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.3 | 1.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 1.3 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.3 | 1.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 2.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 1.2 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.2 | 2.9 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.6 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 0.8 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.5 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.4 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.1 | 2.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.5 | GO:0070476 | RNA (guanine-N7)-methylation(GO:0036265) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 1.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 0.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.7 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 2.1 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.1 | 0.5 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.4 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.7 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.2 | GO:1903414 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.6 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 2.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.2 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.1 | 2.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.8 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 1.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 1.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 1.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 1.8 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.7 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 1.5 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.3 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 3.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 2.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 2.7 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 22.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 7.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.9 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.5 | 24.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.3 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 1.3 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.3 | 2.9 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 1.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 2.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 2.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 1.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 1.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 0.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.5 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 0.2 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.1 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.6 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.8 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 2.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 26.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |