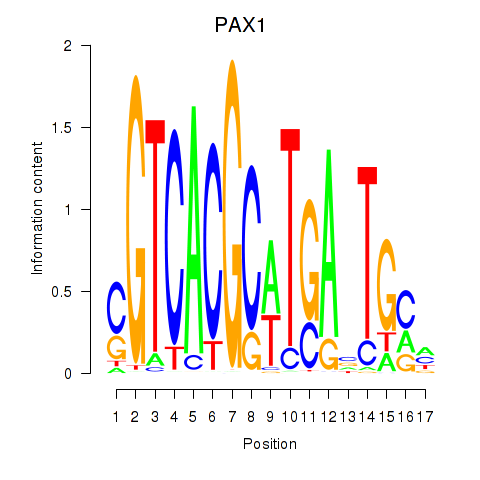
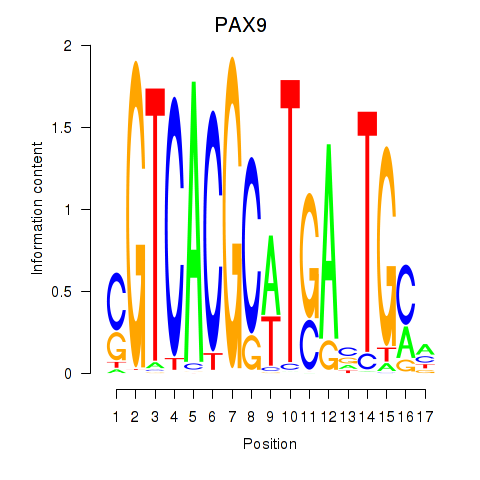
|
chr22_+_22895368
Show fit
|
3.01 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1
|
|
chr4_+_122826824
Show fit
|
2.05 |
ENST00000608478.1
ENST00000644866.2
|
FGF2
|
fibroblast growth factor 2
|
|
chr8_-_23404076
Show fit
|
2.02 |
ENST00000524168.1
ENST00000389131.8
ENST00000523833.2
ENST00000519243.1
|
LOXL2
|
lysyl oxidase like 2
|
|
chr2_+_118942290
Show fit
|
2.00 |
ENST00000412481.1
|
MARCO
|
macrophage receptor with collagenous structure
|
|
chr4_+_122826679
Show fit
|
1.98 |
ENST00000264498.8
|
FGF2
|
fibroblast growth factor 2
|
|
chr8_-_23425282
Show fit
|
1.63 |
ENST00000524144.5
|
LOXL2
|
lysyl oxidase like 2
|
|
chr10_+_71319249
Show fit
|
1.37 |
ENST00000373189.6
ENST00000479577.2
|
SLC29A3
|
solute carrier family 29 member 3
|
|
chr8_-_23425307
Show fit
|
1.37 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase like 2
|
|
chr14_-_68794597
Show fit
|
1.35 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein like 1
|
|
chr18_-_50287570
Show fit
|
1.17 |
ENST00000586837.1
ENST00000412036.6
ENST00000589940.5
ENST00000587396.1
ENST00000591474.5
ENST00000285106.11
|
CXXC1
|
CXXC finger protein 1
|
|
chr10_-_43409160
Show fit
|
1.14 |
ENST00000337970.7
ENST00000682386.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr8_-_100952918
Show fit
|
1.01 |
ENST00000395957.6
ENST00000395948.6
ENST00000457309.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
|
|
chr2_-_162074182
Show fit
|
0.97 |
ENST00000360534.8
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr2_-_162074394
Show fit
|
0.95 |
ENST00000676810.1
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr8_-_100953331
Show fit
|
0.92 |
ENST00000353245.7
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
|
|
chr18_-_49492305
Show fit
|
0.88 |
ENST00000615479.4
ENST00000583637.5
ENST00000618613.5
ENST00000615760.4
ENST00000578528.1
ENST00000578532.5
ENST00000580387.5
ENST00000579248.5
ENST00000580261.6
ENST00000581373.5
ENST00000618619.4
ENST00000617346.4
ENST00000583036.5
ENST00000332968.11
|
RPL17
RPL17-C18orf32
|
ribosomal protein L17
RPL17-C18orf32 readthrough
|
|
chr8_-_100952876
Show fit
|
0.87 |
ENST00000437293.5
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
|
|
chr10_+_30434602
Show fit
|
0.81 |
ENST00000413724.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr10_+_30434021
Show fit
|
0.79 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr3_+_133805789
Show fit
|
0.73 |
ENST00000678299.1
|
SRPRB
|
SRP receptor subunit beta
|
|
chr2_-_162074050
Show fit
|
0.72 |
ENST00000676768.1
|
DPP4
|
dipeptidyl peptidase 4
|
|
chr10_+_30434176
Show fit
|
0.64 |
ENST00000263056.6
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr8_-_120811988
Show fit
|
0.64 |
ENST00000517992.2
|
SNTB1
|
syntrophin beta 1
|
|
chr4_+_74365136
Show fit
|
0.61 |
ENST00000244869.3
|
EREG
|
epiregulin
|
|
chr18_-_49491586
Show fit
|
0.61 |
ENST00000584895.5
ENST00000580210.5
ENST00000579408.5
|
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr14_-_91417805
Show fit
|
0.60 |
ENST00000389857.11
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr17_+_39200275
Show fit
|
0.58 |
ENST00000225430.9
|
RPL19
|
ribosomal protein L19
|
|
chr6_-_137173644
Show fit
|
0.58 |
ENST00000296980.7
ENST00000349184.8
ENST00000339602.3
|
IL22RA2
|
interleukin 22 receptor subunit alpha 2
|
|
chr17_+_39200302
Show fit
|
0.57 |
ENST00000579374.5
|
RPL19
|
ribosomal protein L19
|
|
chr17_+_39200334
Show fit
|
0.56 |
ENST00000579260.5
ENST00000582193.5
|
RPL19
|
ribosomal protein L19
|
|
chr10_+_30434116
Show fit
|
0.54 |
ENST00000415139.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr17_+_39200507
Show fit
|
0.52 |
ENST00000678573.1
|
RPL19
|
ribosomal protein L19
|
|
chr11_-_126268810
Show fit
|
0.49 |
ENST00000332118.11
|
SRPRA
|
SRP receptor subunit alpha
|
|
chr14_+_64388296
Show fit
|
0.39 |
ENST00000554739.5
ENST00000554768.6
ENST00000652179.1
ENST00000652337.1
ENST00000557370.3
|
MTHFD1
|
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1
|
|
chr2_-_174847765
Show fit
|
0.36 |
ENST00000443238.6
|
CHN1
|
chimerin 1
|
|
chr12_+_57434778
Show fit
|
0.33 |
ENST00000309668.3
|
INHBC
|
inhibin subunit beta C
|
|
chr3_+_195720974
Show fit
|
0.33 |
ENST00000447234.7
ENST00000320736.10
|
MUC20
|
mucin 20, cell surface associated
|
|
chr9_-_122228845
Show fit
|
0.32 |
ENST00000394319.8
ENST00000340587.7
|
LHX6
|
LIM homeobox 6
|
|
chr11_+_61755372
Show fit
|
0.32 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor
|
|
chr3_-_57079287
Show fit
|
0.30 |
ENST00000338458.8
ENST00000468727.5
|
ARHGEF3
|
Rho guanine nucleotide exchange factor 3
|
|
chr11_-_126268913
Show fit
|
0.28 |
ENST00000532259.1
|
SRPRA
|
SRP receptor subunit alpha
|
|
chr22_-_17773976
Show fit
|
0.24 |
ENST00000317361.11
|
BID
|
BH3 interacting domain death agonist
|
|
chr6_+_132570322
Show fit
|
0.24 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6
|
|
chrX_-_154479253
Show fit
|
0.23 |
ENST00000357360.4
|
LAGE3
|
L antigen family member 3
|
|
chr17_+_78378633
Show fit
|
0.23 |
ENST00000262764.11
ENST00000589689.5
ENST00000592043.5
ENST00000587356.1
|
PGS1
|
phosphatidylglycerophosphate synthase 1
|
|
chr17_+_41812974
Show fit
|
0.21 |
ENST00000321562.9
|
FKBP10
|
FKBP prolyl isomerase 10
|
|
chr17_-_3557798
Show fit
|
0.21 |
ENST00000301365.8
ENST00000572519.1
ENST00000576742.6
|
TRPV3
|
transient receptor potential cation channel subfamily V member 3
|
|
chr17_-_35487831
Show fit
|
0.19 |
ENST00000260908.12
|
ENSG00000205045.11
|
schlafen family member 12 like
|
|
chr2_-_174847250
Show fit
|
0.19 |
ENST00000409089.7
|
CHN1
|
chimerin 1
|
|
chr1_+_26432299
Show fit
|
0.18 |
ENST00000427245.6
ENST00000236342.12
ENST00000525682.6
ENST00000526219.5
ENST00000374185.7
ENST00000360009.6
ENST00000533087.5
ENST00000531312.5
ENST00000525165.5
ENST00000525326.5
ENST00000525546.5
ENST00000436153.6
ENST00000530781.5
|
DHDDS
|
dehydrodolichyl diphosphate synthase subunit
|
|
chr17_-_35487739
Show fit
|
0.17 |
ENST00000628453.3
|
ENSG00000205045.11
|
schlafen family member 12 like
|
|
chr12_+_57745017
Show fit
|
0.16 |
ENST00000547992.5
ENST00000552816.5
ENST00000257910.8
ENST00000547472.5
|
TSPAN31
|
tetraspanin 31
|
|
chr12_+_131929219
Show fit
|
0.15 |
ENST00000322060.9
|
PUS1
|
pseudouridine synthase 1
|
|
chr7_-_144835981
Show fit
|
0.14 |
ENST00000360057.7
ENST00000378099.7
ENST00000639328.1
|
TPK1
|
thiamin pyrophosphokinase 1
|
|
chr2_+_68643579
Show fit
|
0.14 |
ENST00000303786.5
|
PROKR1
|
prokineticin receptor 1
|
|
chr19_-_38847423
Show fit
|
0.13 |
ENST00000647557.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr12_+_131929194
Show fit
|
0.12 |
ENST00000443358.6
|
PUS1
|
pseudouridine synthase 1
|
|
chr1_-_1574824
Show fit
|
0.11 |
ENST00000291386.4
|
SSU72
|
SSU72 homolog, RNA polymerase II CTD phosphatase
|
|
chr17_+_41812673
Show fit
|
0.11 |
ENST00000585664.5
ENST00000585922.5
ENST00000429461.5
|
FKBP10
|
FKBP prolyl isomerase 10
|
|
chrX_-_149540900
Show fit
|
0.10 |
ENST00000608355.1
ENST00000651111.1
|
LINC00893
ENSG00000241489.8
|
long intergenic non-protein coding RNA 893
novel protein
|
|
chr4_-_170090153
Show fit
|
0.10 |
ENST00000509167.5
ENST00000353187.6
ENST00000507375.5
ENST00000515480.5
|
AADAT
|
aminoadipate aminotransferase
|
|
chr7_+_142770960
Show fit
|
0.10 |
ENST00000632805.1
ENST00000633969.1
ENST00000539842.6
|
PRSS2
|
serine protease 2
|
|
chr6_+_34889228
Show fit
|
0.09 |
ENST00000360359.5
ENST00000649117.1
ENST00000650178.1
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr3_-_57079253
Show fit
|
0.09 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor 3
|
|
chr1_+_26432803
Show fit
|
0.08 |
ENST00000430232.5
|
DHDDS
|
dehydrodolichyl diphosphate synthase subunit
|
|
chr2_+_241809065
Show fit
|
0.07 |
ENST00000415936.5
|
NEU4
|
neuraminidase 4
|
|
chr1_+_44674688
Show fit
|
0.07 |
ENST00000418644.5
ENST00000458657.6
ENST00000535358.6
ENST00000441519.5
ENST00000445071.5
|
ARMH1
|
armadillo like helical domain containing 1
|
|
chr1_-_152115443
Show fit
|
0.07 |
ENST00000614923.1
|
TCHH
|
trichohyalin
|
|
chr9_+_33750669
Show fit
|
0.04 |
ENST00000361005.10
ENST00000342836.9
ENST00000429677.8
|
PRSS3
|
serine protease 3
|
|
chrX_-_132957245
Show fit
|
0.04 |
ENST00000640529.1
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr6_-_170584581
Show fit
|
0.03 |
ENST00000392090.6
ENST00000542896.5
ENST00000453163.6
ENST00000537445.5
ENST00000541970.6
|
PDCD2
|
programmed cell death 2
|
|
chr3_+_31532901
Show fit
|
0.02 |
ENST00000295770.4
|
STT3B
|
STT3 oligosaccharyltransferase complex catalytic subunit B
|
|
chr3_+_195720867
Show fit
|
0.02 |
ENST00000436408.6
|
MUC20
|
mucin 20, cell surface associated
|
|
chr2_-_39187707
Show fit
|
0.01 |
ENST00000451199.5
|
CDKL4
|
cyclin dependent kinase like 4
|