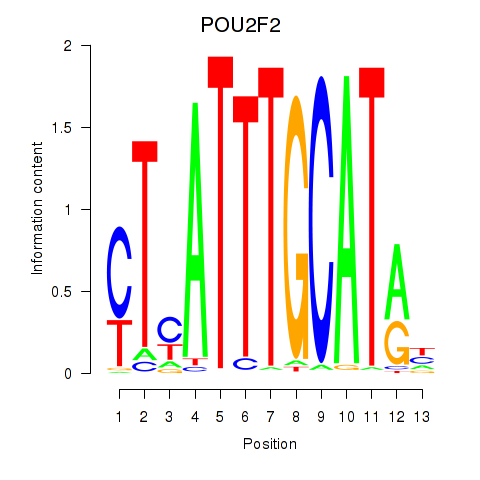
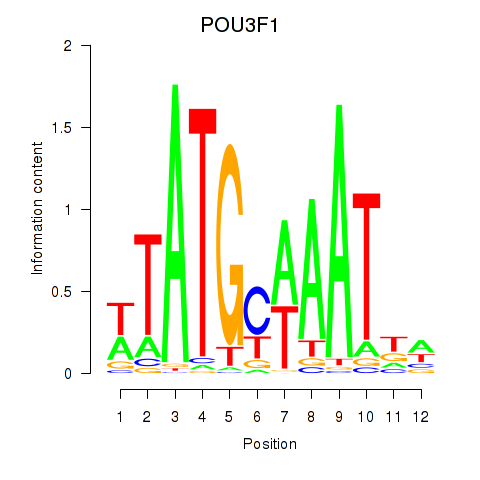
Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for POU2F2_POU3F1
Z-value: 3.54
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.21 | POU2F2 |
|
POU3F1
|
ENSG00000185668.8 | POU3F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F2 | hg38_v1_chr19_-_42132391_42132455 | 0.67 | 2.9e-05 | Click! |
| POU3F1 | hg38_v1_chr1_-_38046785_38046802 | -0.47 | 6.9e-03 | Click! |
Activity profile of POU2F2_POU3F1 motif
Sorted Z-values of POU2F2_POU3F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F2_POU3F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106117159 | 46.05 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr14_-_106538331 | 44.61 |
ENST00000390624.3
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr14_-_106235582 | 41.45 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr14_-_106154113 | 36.09 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr14_-_106803221 | 33.61 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr14_-_106360320 | 30.82 |
ENST00000390615.2
|
IGHV3-33
|
immunoglobulin heavy variable 3-33 |
| chr14_-_106715166 | 30.59 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69 |
| chr14_-_106470788 | 30.42 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr2_-_89040745 | 30.28 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr14_-_106062670 | 30.23 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr2_+_89913982 | 28.80 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr14_-_106335613 | 27.44 |
ENST00000603660.1
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr22_+_22098683 | 27.42 |
ENST00000390283.2
|
IGLV8-61
|
immunoglobulin lambda variable 8-61 |
| chr16_+_32066065 | 27.40 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr14_-_106675544 | 27.38 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr14_-_106211453 | 26.84 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr22_+_22811737 | 25.19 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr2_-_89268506 | 25.08 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr14_-_106557465 | 25.01 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr14_-_106088573 | 23.37 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr22_+_22686724 | 23.37 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr14_-_106811131 | 23.34 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr22_+_22357739 | 23.03 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr2_+_90100235 | 21.87 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr2_+_90154073 | 21.63 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr14_-_106791226 | 21.60 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr14_-_106593319 | 21.25 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr16_+_33009175 | 20.85 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr2_+_90159840 | 20.42 |
ENST00000377032.5
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr16_+_33827140 | 20.35 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr2_-_88992903 | 20.00 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr14_-_106108453 | 19.70 |
ENST00000632950.2
|
IGHV5-10-1
|
immunoglobulin heavy variable 5-10-1 |
| chrX_-_48919015 | 19.59 |
ENST00000376509.4
|
PIM2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr14_-_106130061 | 19.06 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr16_+_32995048 | 18.61 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr2_+_90082635 | 18.30 |
ENST00000483379.1
|
IGKV1D-17
|
immunoglobulin kappa variable 1D-17 |
| chr2_+_88897230 | 18.27 |
ENST00000390244.2
|
IGKV5-2
|
immunoglobulin kappa variable 5-2 |
| chr14_-_106165730 | 17.74 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr14_-_106269133 | 17.73 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr22_+_22880706 | 17.04 |
ENST00000390319.2
|
IGLV3-1
|
immunoglobulin lambda variable 3-1 |
| chr14_-_106658251 | 16.64 |
ENST00000454421.2
|
IGHV3-64
|
immunoglobulin heavy variable 3-64 |
| chr2_-_89100352 | 15.28 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr22_+_22668286 | 15.20 |
ENST00000390304.2
|
IGLV3-27
|
immunoglobulin lambda variable 3-27 |
| chr14_-_106762576 | 15.14 |
ENST00000624687.1
|
IGHV1-69D
|
immunoglobulin heavy variable 1-69D |
| chr2_-_88947820 | 15.10 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr2_+_90172802 | 14.34 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr2_-_89117844 | 13.99 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr2_-_88966767 | 13.97 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr2_+_90038848 | 13.73 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr22_+_22822658 | 13.63 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr2_-_89245596 | 13.40 |
ENST00000468494.1
|
IGKV2-30
|
immunoglobulin kappa variable 2-30 |
| chr16_-_33845229 | 13.33 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr2_+_89862438 | 13.32 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr2_+_90234809 | 12.96 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr22_+_22758698 | 12.67 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr22_+_22030934 | 12.66 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr22_+_22818994 | 12.64 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 |
| chr22_+_22747383 | 12.57 |
ENST00000390311.3
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chr4_-_70666492 | 12.50 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr14_-_106374129 | 11.96 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34 |
| chr14_-_106389858 | 11.77 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr2_+_89936859 | 11.46 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr4_-_70666884 | 11.31 |
ENST00000510614.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr4_-_70666961 | 11.26 |
ENST00000510437.5
|
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr1_+_206557157 | 11.21 |
ENST00000577571.5
|
RASSF5
|
Ras association domain family member 5 |
| chr22_+_22734577 | 11.06 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr22_+_22431949 | 11.02 |
ENST00000390301.3
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr14_-_106277039 | 10.89 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr16_+_23835946 | 10.76 |
ENST00000321728.12
ENST00000643927.1 |
PRKCB
|
protein kinase C beta |
| chr2_+_113406368 | 10.73 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr2_+_90209873 | 10.61 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr22_+_22697789 | 10.58 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr22_+_22720615 | 10.50 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr2_+_89884740 | 10.12 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr2_+_90114838 | 10.04 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chrX_+_12975216 | 9.95 |
ENST00000380635.5
|
TMSB4X
|
thymosin beta 4 X-linked |
| chr2_-_88979016 | 9.89 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr8_+_55879818 | 9.43 |
ENST00000520220.6
ENST00000519728.6 |
LYN
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr2_-_89177160 | 9.24 |
ENST00000484817.1
|
IGKV2-24
|
immunoglobulin kappa variable 2-24 |
| chr22_+_22409755 | 9.07 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chrX_+_12975083 | 8.99 |
ENST00000451311.7
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4 X-linked |
| chr2_-_89320146 | 8.98 |
ENST00000498574.1
|
IGKV1-39
|
immunoglobulin kappa variable 1-39 |
| chr2_-_89297785 | 8.90 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr15_-_19988117 | 8.56 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr21_-_10649835 | 8.54 |
ENST00000622028.1
|
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr8_-_133102477 | 8.37 |
ENST00000522119.5
ENST00000523610.5 ENST00000338087.10 ENST00000521302.5 ENST00000519558.5 ENST00000519747.5 ENST00000517648.5 |
SLA
|
Src like adaptor |
| chr6_-_27146841 | 8.16 |
ENST00000356950.2
|
H2BC12
|
H2B clustered histone 12 |
| chr1_-_149842736 | 7.90 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr14_-_106025628 | 7.49 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr6_-_31592952 | 7.39 |
ENST00000376073.8
ENST00000376072.7 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr1_+_149851053 | 7.28 |
ENST00000607355.2
|
H2AC19
|
H2A clustered histone 19 |
| chr2_+_90004792 | 7.27 |
ENST00000462693.1
|
IGKV2D-24
|
immunoglobulin kappa variable 2D-24 (non-functional) |
| chr1_+_149886906 | 7.16 |
ENST00000331380.4
|
H2AC20
|
H2A clustered histone 20 |
| chr7_-_142813394 | 7.10 |
ENST00000417977.2
|
TRBV30
|
T cell receptor beta variable 30 |
| chr2_+_90021567 | 7.09 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr1_+_116754422 | 7.04 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr22_-_23754376 | 6.90 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr8_-_133102623 | 6.78 |
ENST00000524345.5
|
SLA
|
Src like adaptor |
| chr2_+_89851723 | 6.63 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr4_-_25863537 | 6.59 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr8_-_133102874 | 6.25 |
ENST00000395352.7
|
SLA
|
Src like adaptor |
| chr5_-_141651376 | 6.15 |
ENST00000522783.5
ENST00000519800.1 ENST00000435817.7 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr2_-_89160329 | 6.00 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr12_-_31326111 | 5.79 |
ENST00000539409.5
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr16_-_88686453 | 5.63 |
ENST00000332281.6
|
SNAI3
|
snail family transcriptional repressor 3 |
| chr14_-_106737547 | 5.60 |
ENST00000632209.1
|
IGHV1-69-2
|
immunoglobulin heavy variable 1-69-2 |
| chr14_-_106005574 | 5.29 |
ENST00000390595.3
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr6_-_31592992 | 5.15 |
ENST00000340027.10
|
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr7_+_98211431 | 5.12 |
ENST00000609256.2
|
BHLHA15
|
basic helix-loop-helix family member a15 |
| chr11_-_5227063 | 5.08 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr3_+_151873634 | 5.08 |
ENST00000362032.6
|
SUCNR1
|
succinate receptor 1 |
| chr3_+_98531965 | 5.05 |
ENST00000284311.5
|
GPR15
|
G protein-coupled receptor 15 |
| chr16_+_28931942 | 5.02 |
ENST00000324662.8
ENST00000538922.8 |
CD19
|
CD19 molecule |
| chr12_-_31326142 | 4.96 |
ENST00000337682.9
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr12_+_121626493 | 4.86 |
ENST00000617316.2
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr19_+_3572927 | 4.81 |
ENST00000333651.11
ENST00000417382.5 |
HMG20B
|
high mobility group 20B |
| chr19_-_18397596 | 4.69 |
ENST00000595840.1
ENST00000339007.4 |
LRRC25
|
leucine rich repeat containing 25 |
| chr4_-_107283746 | 4.67 |
ENST00000510463.1
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr17_-_49764123 | 4.67 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr5_-_67196791 | 4.67 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr6_-_26216673 | 4.65 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr14_-_106507476 | 4.64 |
ENST00000390621.3
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr4_-_107283689 | 4.61 |
ENST00000513208.5
|
DKK2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr15_-_19965101 | 4.53 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr22_+_22395005 | 4.51 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr1_+_207496229 | 4.47 |
ENST00000367051.6
ENST00000367053.6 ENST00000367052.6 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr6_-_149484965 | 4.30 |
ENST00000409806.8
|
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr19_-_40425982 | 4.26 |
ENST00000357949.5
|
SERTAD1
|
SERTA domain containing 1 |
| chr6_+_37170133 | 4.25 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr4_+_40192949 | 4.22 |
ENST00000507851.5
ENST00000615577.4 ENST00000613272.4 |
RHOH
|
ras homolog family member H |
| chr6_+_26124161 | 4.17 |
ENST00000377791.4
ENST00000602637.1 |
H2AC6
|
H2A clustered histone 6 |
| chr17_+_40015428 | 4.09 |
ENST00000394149.8
ENST00000225474.6 ENST00000331769.6 ENST00000394148.7 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 |
| chr19_+_17747698 | 4.07 |
ENST00000594202.5
ENST00000252771.11 |
FCHO1
|
FCH and mu domain containing endocytic adaptor 1 |
| chr19_+_17747737 | 4.07 |
ENST00000600676.5
ENST00000600209.5 ENST00000596309.5 ENST00000598539.5 ENST00000597474.5 ENST00000593385.5 ENST00000598067.5 ENST00000593833.5 |
FCHO1
|
FCH and mu domain containing endocytic adaptor 1 |
| chr1_+_207496147 | 3.96 |
ENST00000400960.7
ENST00000367049.9 |
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr22_+_22427517 | 3.91 |
ENST00000390300.2
|
IGLV5-37
|
immunoglobulin lambda variable 5-37 |
| chr3_+_63652663 | 3.88 |
ENST00000343837.8
ENST00000469440.5 |
SNTN
|
sentan, cilia apical structure protein |
| chr19_-_1132208 | 3.87 |
ENST00000438103.6
|
SBNO2
|
strawberry notch homolog 2 |
| chr14_-_106875069 | 3.83 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr8_-_133102643 | 3.75 |
ENST00000519341.5
|
SLA
|
Src like adaptor |
| chr3_-_112641128 | 3.73 |
ENST00000206423.8
|
CCDC80
|
coiled-coil domain containing 80 |
| chr7_+_50308672 | 3.71 |
ENST00000439701.2
ENST00000438033.5 ENST00000492782.6 |
IKZF1
|
IKAROS family zinc finger 1 |
| chrX_+_135520616 | 3.70 |
ENST00000370752.4
ENST00000639893.2 |
INTS6L
|
integrator complex subunit 6 like |
| chr7_+_18495723 | 3.65 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr3_-_112641292 | 3.62 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr19_+_3572971 | 3.62 |
ENST00000453933.5
ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr7_-_27143672 | 3.55 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr19_+_35329161 | 3.47 |
ENST00000341773.10
ENST00000600131.5 ENST00000595780.5 ENST00000597916.5 ENST00000593867.5 ENST00000600424.5 ENST00000599811.5 ENST00000536635.6 ENST00000085219.10 ENST00000544992.6 ENST00000419549.6 |
CD22
|
CD22 molecule |
| chr1_+_170663134 | 3.46 |
ENST00000367760.7
|
PRRX1
|
paired related homeobox 1 |
| chr5_-_159099909 | 3.39 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1 |
| chr6_-_127900958 | 3.26 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr19_-_43667666 | 3.22 |
ENST00000593714.5
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr6_-_11778781 | 3.16 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr19_-_46788586 | 3.12 |
ENST00000542575.6
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr22_+_22588155 | 3.11 |
ENST00000390302.3
|
IGLV2-33
|
immunoglobulin lambda variable 2-33 (non-functional) |
| chr17_+_74737211 | 3.10 |
ENST00000392612.7
ENST00000392610.5 |
RAB37
|
RAB37, member RAS oncogene family |
| chr4_-_153789057 | 3.08 |
ENST00000274063.5
|
SFRP2
|
secreted frizzled related protein 2 |
| chr7_-_100586119 | 3.07 |
ENST00000310300.11
|
LRCH4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr15_-_55249029 | 3.06 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_-_49665603 | 3.04 |
ENST00000596756.5
|
IRF3
|
interferon regulatory factor 3 |
| chr7_+_18495697 | 3.01 |
ENST00000413380.5
ENST00000430454.5 |
HDAC9
|
histone deacetylase 9 |
| chr14_+_21749163 | 2.99 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr7_+_16753731 | 2.98 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr21_-_34888683 | 2.93 |
ENST00000344691.8
ENST00000358356.9 |
RUNX1
|
RUNX family transcription factor 1 |
| chr19_+_54617122 | 2.89 |
ENST00000396331.5
|
LILRB1
|
leukocyte immunoglobulin like receptor B1 |
| chr19_+_39412650 | 2.86 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr7_-_27113825 | 2.82 |
ENST00000522456.1
|
HOXA3
|
homeobox A3 |
| chr6_+_26251607 | 2.82 |
ENST00000619466.2
|
H2BC9
|
H2B clustered histone 9 |
| chr1_+_66332004 | 2.81 |
ENST00000371045.9
ENST00000531025.5 ENST00000526197.5 |
PDE4B
|
phosphodiesterase 4B |
| chr1_+_206406377 | 2.79 |
ENST00000605476.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr4_-_39032922 | 2.73 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr12_-_98644733 | 2.73 |
ENST00000299157.5
ENST00000393042.3 |
IKBIP
|
IKBKB interacting protein |
| chr16_+_33802683 | 2.73 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr12_-_31326171 | 2.72 |
ENST00000542983.1
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr17_+_74737238 | 2.67 |
ENST00000392613.10
ENST00000613645.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr16_-_88941801 | 2.67 |
ENST00000569464.5
ENST00000569443.5 |
CBFA2T3
|
CBFA2/RUNX1 partner transcriptional co-repressor 3 |
| chr10_-_62236112 | 2.65 |
ENST00000315289.6
|
RTKN2
|
rhotekin 2 |
| chr12_+_112906949 | 2.64 |
ENST00000679971.1
ENST00000675868.2 ENST00000550883.2 ENST00000553152.2 ENST00000202917.10 ENST00000679467.1 ENST00000680659.1 ENST00000540589.3 ENST00000552526.2 ENST00000681228.1 ENST00000680934.1 ENST00000681700.1 ENST00000679987.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr18_-_55322334 | 2.54 |
ENST00000630720.3
|
TCF4
|
transcription factor 4 |
| chr1_+_156150008 | 2.53 |
ENST00000355014.6
|
SEMA4A
|
semaphorin 4A |
| chr17_+_40341388 | 2.50 |
ENST00000394086.7
|
RARA
|
retinoic acid receptor alpha |
| chr19_+_3572777 | 2.48 |
ENST00000416526.5
|
HMG20B
|
high mobility group 20B |
| chr2_-_112836702 | 2.47 |
ENST00000416750.1
ENST00000263341.7 ENST00000418817.5 |
IL1B
|
interleukin 1 beta |
| chr1_+_207496268 | 2.46 |
ENST00000529814.1
|
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr1_+_23691742 | 2.42 |
ENST00000374550.8
ENST00000643754.2 |
RPL11
|
ribosomal protein L11 |
| chr4_+_26320975 | 2.41 |
ENST00000509158.6
ENST00000681856.1 ENST00000680140.1 ENST00000355476.8 ENST00000680511.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr21_+_38272410 | 2.39 |
ENST00000398934.5
ENST00000398930.5 |
KCNJ15
|
potassium inwardly rectifying channel subfamily J member 15 |
| chr6_+_26158115 | 2.35 |
ENST00000377777.5
ENST00000289316.2 |
H2BC5
|
H2B clustered histone 5 |
| chr2_-_174598206 | 2.34 |
ENST00000392546.6
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr4_+_123399488 | 2.31 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1 |
| chr5_-_179618032 | 2.28 |
ENST00000519033.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr17_+_14301069 | 2.27 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr19_-_18281612 | 2.22 |
ENST00000252818.5
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr6_-_73521783 | 2.21 |
ENST00000331523.7
ENST00000356303.7 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr6_-_53148822 | 2.19 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr12_-_51270175 | 2.18 |
ENST00000604188.1
ENST00000398453.7 |
SMAGP
|
small cell adhesion glycoprotein |
| chr12_-_51270274 | 2.14 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr7_+_80646436 | 2.14 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr6_+_27808163 | 2.14 |
ENST00000358739.4
|
H2AC13
|
H2A clustered histone 13 |
| chr16_+_56935371 | 2.14 |
ENST00000568358.1
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 1149.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 3.9 | 35.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 3.1 | 9.4 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 2.5 | 2.5 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 2.4 | 18.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 2.1 | 12.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 1.5 | 10.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.3 | 5.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 1.2 | 285.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 1.1 | 3.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 1.0 | 3.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 1.0 | 3.9 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.9 | 2.7 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.9 | 3.5 | GO:0060480 | bronchiole development(GO:0060435) lung goblet cell differentiation(GO:0060480) |
| 0.9 | 7.0 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.8 | 2.5 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.8 | 3.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.8 | 4.7 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.7 | 2.9 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.7 | 2.0 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.7 | 1.3 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.7 | 2.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.6 | 1.3 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.6 | 2.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.6 | 1.8 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.6 | 2.8 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.5 | 2.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.5 | 1.9 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.5 | 2.3 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.5 | 3.2 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.4 | 8.5 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.4 | 3.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.4 | 1.3 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.4 | 2.5 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.4 | 6.1 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.4 | 2.8 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.4 | 6.2 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.4 | 1.1 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 0.4 | 4.0 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.4 | 0.7 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 2.6 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.3 | 2.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 2.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 10.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 7.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.3 | 5.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 0.9 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 7.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 4.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 3.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 2.0 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.2 | 1.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 3.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.2 | 1.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 2.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 1.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 8.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 4.0 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 1.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 0.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 2.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 25.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 19.6 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 4.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 2.0 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 4.9 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 3.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 2.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 5.0 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.1 | 0.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 2.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 2.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 1.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.4 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 0.7 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.1 | 1.4 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 1.9 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 2.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 1.7 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 5.1 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 2.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.5 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 5.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 32.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.4 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 | 2.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 1.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 5.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 1.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.0 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 0.9 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.4 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 2.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 1.0 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 1.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.9 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 5.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 7.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 3.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 9.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 1.2 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 1.5 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 2.3 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.0 | 5.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 7.8 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.6 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 2.0 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 2.8 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 1.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 2.0 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 35.1 | GO:0071757 | hexameric IgM immunoglobulin complex(GO:0071757) |
| 8.7 | 478.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.6 | 9.4 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.3 | 5.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.0 | 213.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 1.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 4.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 0.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 1.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 2.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 2.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 3.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 7.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 18.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 8.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.7 | GO:0043293 | apoptosome(GO:0043293) |
| 0.2 | 3.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 2.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 1.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 344.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 7.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 4.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.8 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 7.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 6.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 17.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 9.0 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 5.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.9 | GO:0098560 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 1.9 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 9.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 13.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 6.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 247.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 4.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 513.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 4.3 | 933.4 | GO:0003823 | antigen binding(GO:0003823) |
| 3.6 | 10.9 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 2.7 | 10.8 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.3 | 10.5 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.7 | 5.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.7 | 2.9 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.7 | 2.0 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.5 | 3.2 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.5 | 9.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 8.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.5 | 1.4 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.5 | 4.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 2.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.4 | 2.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 4.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 4.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 5.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 19.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 3.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 2.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 2.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 2.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 3.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 23.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 5.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 4.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.2 | 7.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 4.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 7.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 7.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 1.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 1.3 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.2 | 1.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 11.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.9 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 2.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 2.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 3.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 2.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 4.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 2.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 2.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 2.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 3.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 3.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0050693 | DBD domain binding(GO:0050692) LBD domain binding(GO:0050693) |
| 0.0 | 2.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 13.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 3.7 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.7 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 4.1 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 20.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 3.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 16.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 10.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 12.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 8.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 7.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 11.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 4.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 7.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 11.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 2.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 4.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 19.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 7.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 4.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 5.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 11.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 5.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 7.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 2.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 7.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |