Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for POU6F2
Z-value: 1.80
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.21 | POU6F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg38_v1_chr7_+_38977904_38978009 | 0.37 | 3.9e-02 | Click! |
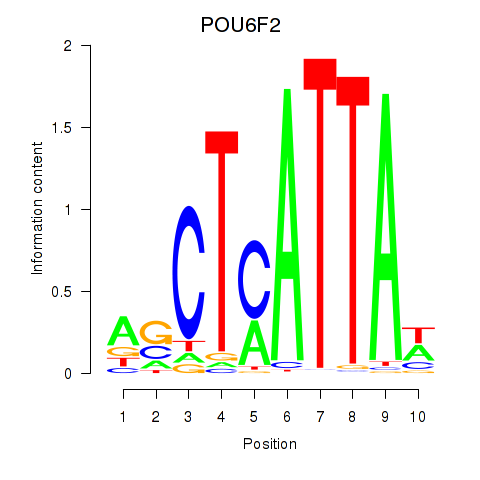
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_43656289 | 5.54 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr3_+_159839847 | 4.34 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_-_73624492 | 4.29 |
ENST00000414749.6
ENST00000429400.6 ENST00000434326.5 ENST00000313375.8 ENST00000354613.5 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein like |
| chr14_+_32329341 | 4.23 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr14_+_32329256 | 3.60 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr10_-_20897288 | 3.48 |
ENST00000377122.9
|
NEBL
|
nebulette |
| chr8_+_91249307 | 3.35 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr15_-_53733103 | 3.03 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr12_-_16605939 | 2.99 |
ENST00000541295.5
ENST00000535535.5 |
LMO3
|
LIM domain only 3 |
| chr10_-_121596117 | 2.90 |
ENST00000351936.11
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr20_+_57561103 | 2.82 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 |
| chr4_+_109912877 | 2.76 |
ENST00000265171.10
ENST00000509793.5 ENST00000652245.1 |
EGF
|
epidermal growth factor |
| chr11_+_107591077 | 2.75 |
ENST00000531234.5
ENST00000265840.12 |
ELMOD1
|
ELMO domain containing 1 |
| chr12_-_16606102 | 2.74 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3 |
| chr11_-_27472698 | 2.68 |
ENST00000389858.4
ENST00000379214.9 |
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr5_-_97142579 | 2.62 |
ENST00000274382.9
|
LIX1
|
limb and CNS expressed 1 |
| chr12_-_16609869 | 2.49 |
ENST00000534946.5
|
LMO3
|
LIM domain only 3 |
| chr17_-_10518536 | 2.42 |
ENST00000226207.6
|
MYH1
|
myosin heavy chain 1 |
| chr7_-_150302980 | 2.33 |
ENST00000252071.8
|
ACTR3C
|
actin related protein 3C |
| chr3_-_33645433 | 2.19 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_44275457 | 2.09 |
ENST00000611973.4
ENST00000409387.5 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr2_+_44275473 | 2.08 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr20_+_43558968 | 2.06 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chrX_-_13817027 | 2.03 |
ENST00000493677.5
ENST00000355135.6 ENST00000316715.9 |
GPM6B
|
glycoprotein M6B |
| chr11_-_129192198 | 1.96 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr12_-_102950835 | 1.92 |
ENST00000546844.1
|
PAH
|
phenylalanine hydroxylase |
| chr9_-_76906041 | 1.91 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr6_+_63521738 | 1.91 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr2_+_44275491 | 1.87 |
ENST00000410056.7
ENST00000409741.5 ENST00000409229.7 |
SLC3A1
|
solute carrier family 3 member 1 |
| chr10_-_126521439 | 1.77 |
ENST00000284694.11
ENST00000432642.5 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr1_-_204359885 | 1.77 |
ENST00000414478.1
ENST00000272203.8 |
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chr17_+_39628496 | 1.76 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr11_-_129192291 | 1.75 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr11_-_82733850 | 1.74 |
ENST00000329203.5
|
FAM181B
|
family with sequence similarity 181 member B |
| chr3_+_111998739 | 1.71 |
ENST00000393917.6
ENST00000273368.8 |
TAGLN3
|
transgelin 3 |
| chr11_-_26572254 | 1.65 |
ENST00000529533.6
|
MUC15
|
mucin 15, cell surface associated |
| chr18_-_26549402 | 1.58 |
ENST00000408011.7
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr7_-_13989658 | 1.56 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr3_+_111998915 | 1.55 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr12_-_86838867 | 1.53 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chr13_-_109786567 | 1.51 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr8_-_141002072 | 1.50 |
ENST00000517453.5
|
PTK2
|
protein tyrosine kinase 2 |
| chr3_+_111999189 | 1.50 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr8_-_42541042 | 1.49 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 member 2 |
| chr5_+_126371306 | 1.49 |
ENST00000506445.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr3_+_189789734 | 1.48 |
ENST00000437221.5
ENST00000392463.6 ENST00000392461.7 ENST00000449992.5 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr9_+_106863121 | 1.46 |
ENST00000472574.1
ENST00000277225.10 |
ZNF462
|
zinc finger protein 462 |
| chr4_-_22443110 | 1.44 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr6_+_121437378 | 1.43 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr5_+_36606355 | 1.43 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr5_-_135578983 | 1.42 |
ENST00000512158.6
|
CXCL14
|
C-X-C motif chemokine ligand 14 |
| chr3_+_35643621 | 1.40 |
ENST00000419330.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr1_-_159862648 | 1.37 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr11_+_45896541 | 1.35 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr7_-_13988863 | 1.29 |
ENST00000405358.8
|
ETV1
|
ETS variant transcription factor 1 |
| chr5_-_97142753 | 1.28 |
ENST00000512378.1
|
LIX1
|
limb and CNS expressed 1 |
| chrX_-_23907887 | 1.28 |
ENST00000379226.9
|
APOO
|
apolipoprotein O |
| chr12_-_16608183 | 1.25 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr1_+_177170916 | 1.24 |
ENST00000361539.5
|
BRINP2
|
BMP/retinoic acid inducible neural specific 2 |
| chr8_-_42501224 | 1.21 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr8_+_42541128 | 1.19 |
ENST00000438528.7
|
SMIM19
|
small integral membrane protein 19 |
| chrX_-_32412220 | 1.18 |
ENST00000619831.5
|
DMD
|
dystrophin |
| chrX_+_11822423 | 1.17 |
ENST00000656302.1
ENST00000640291.2 |
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr12_-_16608073 | 1.16 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr15_+_67098582 | 1.16 |
ENST00000559092.1
ENST00000560175.5 |
SMAD3
|
SMAD family member 3 |
| chr9_-_76906090 | 1.14 |
ENST00000376718.8
|
PRUNE2
|
prune homolog 2 with BCH domain |
| chr6_-_87095059 | 1.11 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr12_-_99984227 | 1.11 |
ENST00000547776.6
ENST00000547010.5 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr11_-_26572102 | 1.09 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr8_+_76681208 | 1.09 |
ENST00000651372.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr15_+_58410543 | 1.06 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr11_-_26572130 | 1.05 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr7_+_129375643 | 1.05 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr11_-_70826887 | 1.05 |
ENST00000656230.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_-_86360010 | 1.05 |
ENST00000641911.1
ENST00000641072.1 ENST00000359221.8 ENST00000640490.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr5_+_90640718 | 1.05 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr7_+_129368123 | 1.04 |
ENST00000460109.5
ENST00000474594.5 |
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr14_-_36519679 | 1.04 |
ENST00000498187.6
|
NKX2-1
|
NK2 homeobox 1 |
| chr18_+_63702958 | 1.04 |
ENST00000544088.6
|
SERPINB11
|
serpin family B member 11 |
| chr3_+_189789643 | 1.04 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr2_-_2324323 | 1.04 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr12_-_70637405 | 1.03 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr5_-_140346596 | 1.01 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr3_+_111999326 | 1.01 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr13_+_77741331 | 0.99 |
ENST00000462234.5
|
SLAIN1
|
SLAIN motif family member 1 |
| chrX_-_21658324 | 0.97 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr13_+_77741393 | 0.95 |
ENST00000496045.5
|
SLAIN1
|
SLAIN motif family member 1 |
| chr17_+_50165990 | 0.95 |
ENST00000262018.8
ENST00000344627.10 ENST00000682109.1 ENST00000504073.2 |
SGCA
|
sarcoglycan alpha |
| chr5_-_157059109 | 0.95 |
ENST00000523175.6
ENST00000522693.5 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr12_-_16610037 | 0.94 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 |
| chr14_-_26598025 | 0.92 |
ENST00000539517.7
|
NOVA1
|
NOVA alternative splicing regulator 1 |
| chr22_+_20550981 | 0.92 |
ENST00000457322.5
|
MED15
|
mediator complex subunit 15 |
| chr6_+_151325665 | 0.92 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr4_-_99144238 | 0.88 |
ENST00000512499.5
ENST00000504125.1 ENST00000505590.5 ENST00000629236.2 ENST00000508393.5 ENST00000265512.12 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr20_+_43559060 | 0.86 |
ENST00000485914.2
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr1_+_99646025 | 0.85 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr13_-_44161257 | 0.83 |
ENST00000400419.2
|
SMIM2
|
small integral membrane protein 2 |
| chr12_+_15546344 | 0.82 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr12_-_130633825 | 0.82 |
ENST00000541840.2
|
RIMBP2
|
RIMS binding protein 2 |
| chr17_+_7407838 | 0.81 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chrX_-_18672101 | 0.79 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr1_-_47190013 | 0.78 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr2_+_6978624 | 0.78 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr1_-_193059489 | 0.77 |
ENST00000367455.8
ENST00000421683.1 |
UCHL5
|
ubiquitin C-terminal hydrolase L5 |
| chr21_-_34526850 | 0.77 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr1_-_193059296 | 0.76 |
ENST00000367450.7
ENST00000367451.8 ENST00000367454.6 ENST00000367448.5 ENST00000367449.5 |
UCHL5
|
ubiquitin C-terminal hydrolase L5 |
| chr7_+_117020191 | 0.72 |
ENST00000434836.5
ENST00000393443.5 ENST00000465133.5 ENST00000477742.5 ENST00000393444.7 ENST00000393447.8 |
ST7
|
suppression of tumorigenicity 7 |
| chr6_+_50818857 | 0.72 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr1_+_50108856 | 0.71 |
ENST00000650764.1
ENST00000494555.2 ENST00000371824.7 ENST00000371823.8 ENST00000652693.1 |
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr22_+_20551135 | 0.69 |
ENST00000424287.5
ENST00000423862.5 |
MED15
|
mediator complex subunit 15 |
| chr11_+_65787056 | 0.69 |
ENST00000335987.8
|
OVOL1
|
ovo like transcriptional repressor 1 |
| chr7_+_129375830 | 0.69 |
ENST00000466993.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr17_+_75543258 | 0.69 |
ENST00000581713.5
|
LLGL2
|
LLGL scribble cell polarity complex component 2 |
| chr6_-_142946312 | 0.66 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr4_+_112860912 | 0.66 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr6_-_155314444 | 0.65 |
ENST00000367166.5
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr6_-_138545685 | 0.64 |
ENST00000342260.9
|
NHSL1
|
NHS like 1 |
| chr21_-_30492008 | 0.64 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr17_+_12955763 | 0.63 |
ENST00000583608.1
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr1_+_50105666 | 0.61 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr22_+_20551000 | 0.60 |
ENST00000428629.5
|
MED15
|
mediator complex subunit 15 |
| chr4_-_73620391 | 0.60 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr4_+_87650277 | 0.59 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr11_-_93850559 | 0.58 |
ENST00000409977.2
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr10_-_73655984 | 0.58 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like |
| chr4_+_112861053 | 0.58 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr4_-_65670478 | 0.57 |
ENST00000613740.5
ENST00000622150.4 ENST00000511294.1 |
EPHA5
|
EPH receptor A5 |
| chr21_-_31038476 | 0.57 |
ENST00000382822.2
|
KRTAP19-8
|
keratin associated protein 19-8 |
| chr4_+_112860981 | 0.57 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr5_+_132873660 | 0.56 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr4_+_109827963 | 0.54 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr21_-_34526815 | 0.54 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr1_-_153150884 | 0.53 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr5_-_124746630 | 0.53 |
ENST00000513986.2
|
ZNF608
|
zinc finger protein 608 |
| chr5_+_127649018 | 0.52 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr3_-_127822455 | 0.50 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr2_+_28778848 | 0.50 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1 catalytic subunit beta |
| chr6_+_25754699 | 0.49 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr1_-_10964201 | 0.49 |
ENST00000418570.6
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr4_-_73620629 | 0.46 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr22_-_50085414 | 0.46 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr3_+_189789672 | 0.45 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr17_-_41424583 | 0.44 |
ENST00000225550.4
|
KRT37
|
keratin 37 |
| chr7_+_1232865 | 0.42 |
ENST00000316333.9
|
UNCX
|
UNC homeobox |
| chr13_-_44160855 | 0.41 |
ENST00000617002.1
|
SMIM2
|
small integral membrane protein 2 |
| chrX_+_30235894 | 0.41 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr6_+_75620639 | 0.40 |
ENST00000483859.6
|
SENP6
|
SUMO specific peptidase 6 |
| chr19_+_38374549 | 0.40 |
ENST00000620216.4
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr13_+_97960192 | 0.40 |
ENST00000496368.6
ENST00000421861.7 ENST00000357602.7 |
IPO5
|
importin 5 |
| chr12_-_85836372 | 0.37 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr19_+_38374557 | 0.37 |
ENST00000215071.9
|
PSMD8
|
proteasome 26S subunit, non-ATPase 8 |
| chr19_-_7632971 | 0.37 |
ENST00000598935.5
|
PCP2
|
Purkinje cell protein 2 |
| chr1_+_45913647 | 0.35 |
ENST00000674079.1
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr5_+_135579193 | 0.34 |
ENST00000646290.1
|
SLC25A48
|
solute carrier family 25 member 48 |
| chr15_-_37101205 | 0.34 |
ENST00000338564.9
ENST00000558313.5 ENST00000340545.9 |
MEIS2
|
Meis homeobox 2 |
| chr11_-_72112068 | 0.34 |
ENST00000537644.5
ENST00000538919.5 ENST00000539395.1 ENST00000542531.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr1_-_208244375 | 0.34 |
ENST00000367033.4
|
PLXNA2
|
plexin A2 |
| chr2_-_80304700 | 0.33 |
ENST00000295057.4
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr22_-_50085331 | 0.33 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr10_+_18260715 | 0.32 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr22_-_50085378 | 0.31 |
ENST00000442311.1
|
MLC1
|
modulator of VRAC current 1 |
| chr8_+_26293112 | 0.30 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr6_+_50818701 | 0.30 |
ENST00000344788.7
|
TFAP2B
|
transcription factor AP-2 beta |
| chr11_-_72112669 | 0.30 |
ENST00000545944.5
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr7_-_143885356 | 0.30 |
ENST00000485416.1
|
TCAF1
|
TRPM8 channel associated factor 1 |
| chr14_-_20641691 | 0.29 |
ENST00000320704.3
|
OR6S1
|
olfactory receptor family 6 subfamily S member 1 |
| chr6_+_42155399 | 0.28 |
ENST00000623004.2
ENST00000372963.4 ENST00000654459.1 |
GUCA1ANB
GUCA1A
|
GUCA1A neighbor guanylate cyclase activator 1A |
| chr11_-_72112750 | 0.28 |
ENST00000545680.5
ENST00000543587.5 ENST00000538393.5 ENST00000535234.5 ENST00000227618.8 ENST00000535503.5 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr3_+_2511597 | 0.28 |
ENST00000434053.5
|
CNTN4
|
contactin 4 |
| chr6_+_158017048 | 0.26 |
ENST00000638626.1
|
SYNJ2
|
synaptojanin 2 |
| chr13_+_57631735 | 0.26 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr9_-_114806031 | 0.25 |
ENST00000374045.5
|
TNFSF15
|
TNF superfamily member 15 |
| chr3_-_51875597 | 0.24 |
ENST00000446461.2
|
IQCF5
|
IQ motif containing F5 |
| chr10_+_47322450 | 0.24 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr17_-_41397600 | 0.22 |
ENST00000251645.3
|
KRT31
|
keratin 31 |
| chr5_+_141412979 | 0.22 |
ENST00000612503.1
ENST00000398610.3 |
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr19_+_9093179 | 0.21 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chrX_+_23908006 | 0.20 |
ENST00000379211.8
ENST00000648352.1 |
CXorf58
|
chromosome X open reading frame 58 |
| chr1_-_201171545 | 0.19 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr4_-_73620500 | 0.19 |
ENST00000335049.5
|
RASSF6
|
Ras association domain family member 6 |
| chr1_+_45913583 | 0.19 |
ENST00000372008.6
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr1_+_193059448 | 0.18 |
ENST00000432079.5
|
RO60
|
Ro60, Y RNA binding protein |
| chr11_-_31509569 | 0.18 |
ENST00000526776.5
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr11_-_120138104 | 0.18 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr6_+_150721073 | 0.17 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr11_-_36598221 | 0.17 |
ENST00000311485.8
ENST00000527033.5 ENST00000532616.1 ENST00000618712.4 |
RAG2
|
recombination activating 2 |
| chr6_-_33192454 | 0.17 |
ENST00000395194.1
ENST00000341947.7 ENST00000374708.8 |
COL11A2
|
collagen type XI alpha 2 chain |
| chr5_-_141618914 | 0.16 |
ENST00000518047.5
|
DIAPH1
|
diaphanous related formin 1 |
| chr11_-_70826716 | 0.16 |
ENST00000468619.6
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr17_+_18183052 | 0.15 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr2_-_80304274 | 0.15 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr9_-_13165442 | 0.14 |
ENST00000542239.1
ENST00000538841.5 ENST00000433359.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr21_+_32298849 | 0.13 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr1_+_207325629 | 0.13 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr4_+_174283699 | 0.12 |
ENST00000505124.5
|
CEP44
|
centrosomal protein 44 |
| chr17_+_18183803 | 0.11 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr3_+_77039836 | 0.11 |
ENST00000461745.5
|
ROBO2
|
roundabout guidance receptor 2 |
| chr12_-_15662692 | 0.11 |
ENST00000540613.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr15_+_101812202 | 0.10 |
ENST00000332238.5
|
OR4F15
|
olfactory receptor family 4 subfamily F member 15 |
| chr4_-_174283614 | 0.09 |
ENST00000513696.1
ENST00000393674.7 ENST00000503293.5 |
FBXO8
|
F-box protein 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.9 | 2.8 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.9 | 7.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.7 | 3.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.7 | 6.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.6 | 2.8 | GO:1900127 | negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.5 | 2.7 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.5 | 2.9 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.4 | 1.4 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.3 | 2.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 1.0 | GO:0097274 | ammonia homeostasis(GO:0097272) urea homeostasis(GO:0097274) |
| 0.3 | 4.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.3 | 5.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.3 | 2.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.4 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 0.8 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.8 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 3.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 3.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 2.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 2.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.7 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 1.5 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 1.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 1.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 1.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 1.0 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.8 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.6 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 2.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 1.9 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 3.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.3 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.1 | 0.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.5 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 1.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 3.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.0 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.1 | GO:0071397 | regulation of response to osmotic stress(GO:0047484) cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.9 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.5 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 3.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 1.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 2.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 2.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 1.5 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.7 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 1.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.5 | GO:0048665 | neuron fate specification(GO:0048665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 1.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 6.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 4.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 5.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 5.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 6.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.5 | 5.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.8 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.4 | 2.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 8.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.9 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.3 | 2.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 3.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 3.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 1.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.9 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 2.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 3.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 3.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 1.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 1.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 1.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 1.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.9 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 4.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 4.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 3.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 1.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 5.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 2.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.9 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 2.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 3.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 6.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 7.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 4.6 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |