Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
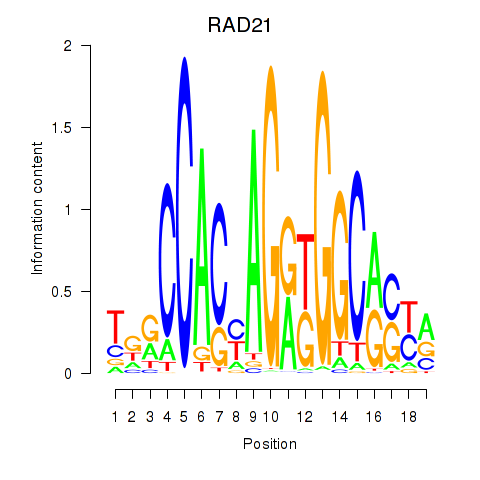
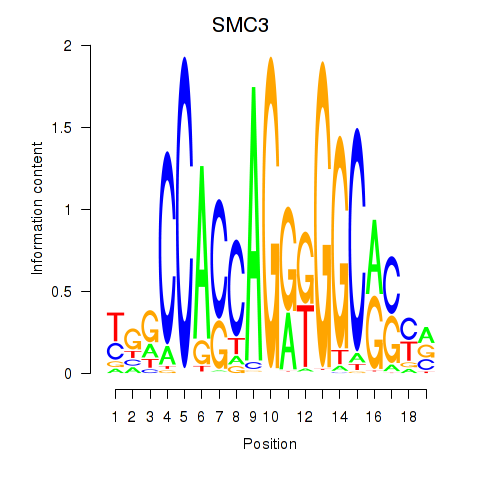
Results for RAD21_SMC3
Z-value: 1.54
Transcription factors associated with RAD21_SMC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RAD21
|
ENSG00000164754.15 | RAD21 |
|
SMC3
|
ENSG00000108055.10 | SMC3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RAD21 | hg38_v1_chr8_-_116874746_116874828 | 0.39 | 2.7e-02 | Click! |
| SMC3 | hg38_v1_chr10_+_110567666_110567765 | 0.29 | 1.0e-01 | Click! |
Activity profile of RAD21_SMC3 motif
Sorted Z-values of RAD21_SMC3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RAD21_SMC3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_22195753 | 3.40 |
ENST00000390285.4
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr7_+_142626642 | 3.38 |
ENST00000390394.3
|
TRBV20-1
|
T cell receptor beta variable 20-1 |
| chr11_+_1876467 | 3.31 |
ENST00000432093.1
|
LSP1
|
lymphocyte specific protein 1 |
| chr22_+_22409755 | 3.23 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr14_+_21868822 | 3.14 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr14_+_21887848 | 3.08 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr22_+_22343185 | 2.96 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr14_+_21723693 | 2.89 |
ENST00000390425.2
|
TRAV3
|
T cell receptor alpha variable 3 |
| chr7_+_142740206 | 2.73 |
ENST00000422143.2
|
TRBV29-1
|
T cell receptor beta variable 29-1 |
| chr22_+_22380766 | 2.68 |
ENST00000390297.3
|
IGLV1-44
|
immunoglobulin lambda variable 1-44 |
| chr22_+_22369601 | 2.67 |
ENST00000390295.3
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 |
| chr19_-_55140922 | 2.53 |
ENST00000589745.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr14_+_21852457 | 2.52 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr22_+_22697789 | 2.47 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr17_+_47733228 | 2.41 |
ENST00000177694.2
|
TBX21
|
T-box transcription factor 21 |
| chr14_+_21978440 | 2.37 |
ENST00000390443.3
|
TRAV8-6
|
T cell receptor alpha variable 8-6 |
| chr16_+_67645100 | 2.34 |
ENST00000334583.11
|
CARMIL2
|
capping protein regulator and myosin 1 linker 2 |
| chr14_+_21918161 | 2.34 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr16_+_67645166 | 2.33 |
ENST00000545661.5
|
CARMIL2
|
capping protein regulator and myosin 1 linker 2 |
| chr14_+_21894433 | 2.31 |
ENST00000390438.2
|
TRAV8-4
|
T cell receptor alpha variable 8-4 |
| chr9_-_131739931 | 2.21 |
ENST00000438647.3
|
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr14_+_21997531 | 2.19 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr19_-_55141889 | 2.19 |
ENST00000593194.5
|
TNNT1
|
troponin T1, slow skeletal type |
| chr14_+_22320128 | 2.17 |
ENST00000390468.1
|
TRAV41
|
T cell receptor alpha variable 41 |
| chr19_+_18007182 | 2.17 |
ENST00000595712.6
|
ARRDC2
|
arrestin domain containing 2 |
| chr9_-_131740056 | 2.15 |
ENST00000372195.5
ENST00000683357.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr6_-_41705813 | 1.98 |
ENST00000419574.6
ENST00000445214.2 |
TFEB
|
transcription factor EB |
| chr7_-_38363476 | 1.98 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr14_+_21749163 | 1.98 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr15_-_64046322 | 1.93 |
ENST00000457488.5
ENST00000612884.4 |
DAPK2
|
death associated protein kinase 2 |
| chr18_+_46173495 | 1.89 |
ENST00000587591.5
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr19_+_49474208 | 1.89 |
ENST00000597551.6
ENST00000596435.5 ENST00000204637.6 ENST00000600429.5 |
FLT3LG
|
fms related receptor tyrosine kinase 3 ligand |
| chr11_+_67266437 | 1.88 |
ENST00000308595.10
ENST00000526285.1 |
GRK2
|
G protein-coupled receptor kinase 2 |
| chrX_-_107777038 | 1.86 |
ENST00000480691.2
ENST00000506081.5 ENST00000514426.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr14_+_22040576 | 1.84 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr11_+_46347526 | 1.81 |
ENST00000456247.6
ENST00000421244.6 ENST00000318201.12 |
DGKZ
|
diacylglycerol kinase zeta |
| chr1_+_151156627 | 1.81 |
ENST00000368910.4
|
TNFAIP8L2
|
TNF alpha induced protein 8 like 2 |
| chr14_+_21841182 | 1.80 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr19_-_19643597 | 1.75 |
ENST00000587205.1
ENST00000203556.9 |
GMIP
|
GEM interacting protein |
| chr17_+_74203582 | 1.73 |
ENST00000439590.6
ENST00000311111.11 ENST00000584577.5 ENST00000534490.5 ENST00000528433.2 ENST00000533498.1 |
RPL38
|
ribosomal protein L38 |
| chr22_+_22357739 | 1.73 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr19_-_19643547 | 1.69 |
ENST00000587238.5
|
GMIP
|
GEM interacting protein |
| chr7_-_38359120 | 1.66 |
ENST00000390346.2
|
TRGV3
|
T cell receptor gamma variable 3 |
| chr2_+_233354474 | 1.65 |
ENST00000264057.7
ENST00000427930.5 ENST00000442524.4 |
DGKD
|
diacylglycerol kinase delta |
| chr22_+_22395005 | 1.61 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr11_-_17389323 | 1.61 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly rectifying channel subfamily J member 11 |
| chr11_+_46347425 | 1.60 |
ENST00000527911.5
|
DGKZ
|
diacylglycerol kinase zeta |
| chr19_-_41353904 | 1.60 |
ENST00000221930.6
|
TGFB1
|
transforming growth factor beta 1 |
| chr10_-_29735787 | 1.54 |
ENST00000375400.7
|
SVIL
|
supervillin |
| chr14_+_80955043 | 1.51 |
ENST00000541158.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr6_-_41940537 | 1.50 |
ENST00000512426.5
|
CCND3
|
cyclin D3 |
| chr14_+_21797272 | 1.48 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr15_-_38564635 | 1.46 |
ENST00000450598.6
ENST00000559830.5 ENST00000558164.5 ENST00000539159.5 ENST00000310803.10 |
RASGRP1
|
RAS guanyl releasing protein 1 |
| chr14_+_22304051 | 1.35 |
ENST00000390466.1
|
TRAV39
|
T cell receptor alpha variable 39 |
| chr17_-_8965674 | 1.35 |
ENST00000447110.6
ENST00000611902.4 ENST00000616147.4 ENST00000623421.3 |
PIK3R5
|
phosphoinositide-3-kinase regulatory subunit 5 |
| chr19_-_6481769 | 1.35 |
ENST00000381480.7
ENST00000543576.5 ENST00000590173.5 |
DENND1C
|
DENN domain containing 1C |
| chr7_-_38354517 | 1.34 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr12_-_120265719 | 1.33 |
ENST00000637617.2
ENST00000267257.11 ENST00000228307.11 ENST00000424649.6 |
PXN
|
paxillin |
| chr15_+_74782069 | 1.33 |
ENST00000220003.14
ENST00000439220.6 |
CSK
|
C-terminal Src kinase |
| chr17_-_9959455 | 1.32 |
ENST00000580865.5
ENST00000583882.5 |
GAS7
|
growth arrest specific 7 |
| chr11_-_64744102 | 1.32 |
ENST00000431822.5
ENST00000394432.8 ENST00000377486.7 |
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr7_+_6589990 | 1.30 |
ENST00000344417.10
|
C7orf26
|
chromosome 7 open reading frame 26 |
| chr10_+_102394488 | 1.28 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2 |
| chr14_+_80955366 | 1.28 |
ENST00000342443.10
|
TSHR
|
thyroid stimulating hormone receptor |
| chr22_+_22431949 | 1.27 |
ENST00000390301.3
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr22_-_21952827 | 1.27 |
ENST00000397495.8
ENST00000263212.10 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F |
| chr17_+_44350437 | 1.26 |
ENST00000586443.1
|
GRN
|
granulin precursor |
| chr19_+_4791710 | 1.25 |
ENST00000269856.5
|
FEM1A
|
fem-1 homolog A |
| chr17_+_27456393 | 1.24 |
ENST00000644974.1
|
KSR1
|
kinase suppressor of ras 1 |
| chr14_+_21965451 | 1.22 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr16_-_69754913 | 1.20 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr19_-_48993300 | 1.19 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr19_-_6481749 | 1.19 |
ENST00000588421.1
|
DENND1C
|
DENN domain containing 1C |
| chr11_+_117232725 | 1.18 |
ENST00000531287.5
ENST00000531452.5 |
RNF214
|
ring finger protein 214 |
| chr5_+_138352674 | 1.18 |
ENST00000314358.10
|
KDM3B
|
lysine demethylase 3B |
| chr19_+_5690255 | 1.18 |
ENST00000582463.5
ENST00000347512.8 ENST00000579446.1 ENST00000394580.2 |
RPL36
|
ribosomal protein L36 |
| chr1_+_28518136 | 1.17 |
ENST00000373832.5
ENST00000373831.7 |
RCC1
|
regulator of chromosome condensation 1 |
| chr14_+_21768482 | 1.14 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr1_+_92832065 | 1.14 |
ENST00000315741.5
|
RPL5
|
ribosomal protein L5 |
| chr6_-_32953017 | 1.14 |
ENST00000395305.7
ENST00000374843.9 ENST00000395303.7 ENST00000429234.1 |
HLA-DMA
ENSG00000248993.1
|
major histocompatibility complex, class II, DM alpha novel protein |
| chr10_-_75109172 | 1.13 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr10_-_29735873 | 1.12 |
ENST00000674490.1
|
SVIL
|
supervillin |
| chr11_-_17389083 | 1.11 |
ENST00000526912.1
|
KCNJ11
|
potassium inwardly rectifying channel subfamily J member 11 |
| chr19_-_48646155 | 1.10 |
ENST00000084798.9
|
CA11
|
carbonic anhydrase 11 |
| chr8_-_27992663 | 1.10 |
ENST00000380385.6
ENST00000354914.8 |
SCARA5
|
scavenger receptor class A member 5 |
| chr20_+_58852710 | 1.09 |
ENST00000676826.2
ENST00000371100.9 |
GNAS
|
GNAS complex locus |
| chr9_+_36136752 | 1.09 |
ENST00000619700.1
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr9_+_36136703 | 1.07 |
ENST00000377960.9
ENST00000377959.5 |
GLIPR2
|
GLI pathogenesis related 2 |
| chr22_-_21938557 | 1.06 |
ENST00000424647.1
ENST00000407142.5 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent 1F |
| chr19_+_49474561 | 1.04 |
ENST00000594009.5
ENST00000595510.1 ENST00000595815.1 |
FLT3LG
ENSG00000273189.1
|
fms related receptor tyrosine kinase 3 ligand novel transcript |
| chr9_+_36136416 | 1.03 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr19_-_18941184 | 1.02 |
ENST00000594794.5
ENST00000392351.8 ENST00000596482.5 |
HOMER3
|
homer scaffold protein 3 |
| chr7_+_6590070 | 1.02 |
ENST00000359073.9
|
C7orf26
|
chromosome 7 open reading frame 26 |
| chr1_+_92832005 | 1.02 |
ENST00000645300.1
|
RPL5
|
ribosomal protein L5 |
| chr17_+_16415553 | 1.00 |
ENST00000338560.12
|
TRPV2
|
transient receptor potential cation channel subfamily V member 2 |
| chr1_+_28518266 | 1.00 |
ENST00000411533.5
|
RCC1
|
regulator of chromosome condensation 1 |
| chr2_-_219160540 | 0.99 |
ENST00000457600.2
|
NHEJ1
|
non-homologous end joining factor 1 |
| chr14_+_55271344 | 0.98 |
ENST00000681400.1
ENST00000679934.1 ENST00000681904.1 ENST00000313833.5 |
FBXO34
|
F-box protein 34 |
| chr16_-_11797208 | 0.97 |
ENST00000571198.5
ENST00000572781.5 ENST00000355758.9 |
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr19_-_35135180 | 0.96 |
ENST00000392225.7
|
LGI4
|
leucine rich repeat LGI family member 4 |
| chr1_-_39901861 | 0.95 |
ENST00000372816.3
ENST00000372815.1 |
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr6_+_106360668 | 0.93 |
ENST00000633556.3
|
CRYBG1
|
crystallin beta-gamma domain containing 1 |
| chr1_-_39901996 | 0.91 |
ENST00000397332.2
|
MYCL
|
MYCL proto-oncogene, bHLH transcription factor |
| chr9_+_126860625 | 0.91 |
ENST00000319119.4
|
ZBTB34
|
zinc finger and BTB domain containing 34 |
| chr1_+_151156659 | 0.91 |
ENST00000602841.5
|
SCNM1
|
sodium channel modifier 1 |
| chr3_+_49803212 | 0.91 |
ENST00000333323.6
|
INKA1
|
inka box actin regulator 1 |
| chr1_+_92832028 | 0.90 |
ENST00000370321.8
ENST00000645119.1 |
RPL5
|
ribosomal protein L5 |
| chr17_-_8383164 | 0.90 |
ENST00000584164.6
ENST00000582556.5 ENST00000648839.1 ENST00000578812.5 ENST00000583011.6 |
RPL26
|
ribosomal protein L26 |
| chr19_+_1941118 | 0.90 |
ENST00000255641.13
|
CSNK1G2
|
casein kinase 1 gamma 2 |
| chr8_-_143430727 | 0.89 |
ENST00000333480.3
|
MAFA
|
MAF bZIP transcription factor A |
| chr19_+_1266653 | 0.88 |
ENST00000586472.5
ENST00000589266.5 |
CIRBP
|
cold inducible RNA binding protein |
| chr19_-_18941117 | 0.88 |
ENST00000600077.5
|
HOMER3
|
homer scaffold protein 3 |
| chr11_-_66336396 | 0.86 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr12_+_109900258 | 0.86 |
ENST00000405876.9
|
TCHP
|
trichoplein keratin filament binding |
| chr12_-_120201099 | 0.86 |
ENST00000551150.5
ENST00000313104.9 ENST00000547191.5 ENST00000546989.5 ENST00000392514.9 ENST00000228306.8 ENST00000550856.5 |
RPLP0
|
ribosomal protein lateral stalk subunit P0 |
| chr14_+_21825453 | 0.86 |
ENST00000390432.2
|
TRAV10
|
T cell receptor alpha variable 10 |
| chr15_+_91853690 | 0.85 |
ENST00000318445.11
|
SLCO3A1
|
solute carrier organic anion transporter family member 3A1 |
| chr11_+_3854527 | 0.82 |
ENST00000525403.5
ENST00000430222.1 |
STIM1
ENSG00000229368.1
|
stromal interaction molecule 1 novel transcript, sense overlapping STIM1 |
| chr14_-_22823386 | 0.81 |
ENST00000554741.5
|
SLC7A7
|
solute carrier family 7 member 7 |
| chr17_-_63701140 | 0.81 |
ENST00000584645.1
|
LIMD2
|
LIM domain containing 2 |
| chr1_-_202160577 | 0.80 |
ENST00000629151.2
ENST00000476061.5 ENST00000464870.5 ENST00000467283.5 ENST00000435759.6 ENST00000486116.5 ENST00000477625.5 |
PTPN7
|
protein tyrosine phosphatase non-receptor type 7 |
| chr5_+_134524305 | 0.80 |
ENST00000431355.2
|
JADE2
|
jade family PHD finger 2 |
| chr11_+_7987314 | 0.79 |
ENST00000531572.2
ENST00000651655.1 |
EIF3F
|
eukaryotic translation initiation factor 3 subunit F |
| chr16_+_69311339 | 0.79 |
ENST00000254950.13
|
VPS4A
|
vacuolar protein sorting 4 homolog A |
| chr8_+_66429003 | 0.79 |
ENST00000320270.4
|
RRS1
|
ribosome biogenesis regulator 1 homolog |
| chr12_-_101830926 | 0.78 |
ENST00000299314.12
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta |
| chr2_-_165203870 | 0.77 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr19_+_55654115 | 0.77 |
ENST00000450554.6
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr17_+_7688427 | 0.77 |
ENST00000396463.7
ENST00000534050.5 |
WRAP53
|
WD repeat containing antisense to TP53 |
| chr12_-_56189548 | 0.76 |
ENST00000347471.8
ENST00000267064.8 ENST00000394023.7 |
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 |
| chr13_-_52450590 | 0.76 |
ENST00000378060.9
|
VPS36
|
vacuolar protein sorting 36 homolog |
| chr2_+_227472132 | 0.76 |
ENST00000409979.6
ENST00000310078.13 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr11_-_18106019 | 0.75 |
ENST00000532452.5
ENST00000530180.1 ENST00000524803.6 ENST00000300013.8 ENST00000529318.5 |
SAAL1
|
serum amyloid A like 1 |
| chr16_-_57536543 | 0.75 |
ENST00000258214.3
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr19_-_41353044 | 0.75 |
ENST00000600196.2
ENST00000677934.1 |
TGFB1
|
transforming growth factor beta 1 |
| chr14_+_22207502 | 0.75 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr14_+_105491226 | 0.74 |
ENST00000451719.5
ENST00000392523.9 ENST00000392522.7 ENST00000354560.10 ENST00000334656.11 ENST00000450383.1 |
TEDC1
|
tubulin epsilon and delta complex 1 |
| chr2_-_231530427 | 0.74 |
ENST00000305141.5
|
NMUR1
|
neuromedin U receptor 1 |
| chr12_+_109900518 | 0.73 |
ENST00000312777.9
ENST00000536408.2 |
TCHP
|
trichoplein keratin filament binding |
| chrX_-_72277235 | 0.73 |
ENST00000316084.10
|
RPS4X
|
ribosomal protein S4 X-linked |
| chr7_-_27180230 | 0.72 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr10_-_75109085 | 0.72 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr7_-_6348906 | 0.72 |
ENST00000313324.9
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220 member A |
| chr3_+_48465811 | 0.71 |
ENST00000433541.1
ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr11_+_117232625 | 0.71 |
ENST00000534428.5
ENST00000300650.9 |
RNF214
|
ring finger protein 214 |
| chr16_+_4624811 | 0.70 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr6_-_41735557 | 0.69 |
ENST00000373033.6
|
TFEB
|
transcription factor EB |
| chrX_-_46759055 | 0.69 |
ENST00000328306.4
ENST00000616978.5 |
SLC9A7
|
solute carrier family 9 member A7 |
| chr22_+_22720615 | 0.69 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr7_+_149147422 | 0.68 |
ENST00000475153.6
|
ZNF398
|
zinc finger protein 398 |
| chr8_-_144787275 | 0.68 |
ENST00000343459.8
ENST00000429371.7 ENST00000534445.1 |
ZNF34
|
zinc finger protein 34 |
| chr17_-_81869934 | 0.68 |
ENST00000580685.5
|
ARHGDIA
|
Rho GDP dissociation inhibitor alpha |
| chr17_+_2055094 | 0.67 |
ENST00000399849.4
ENST00000619757.5 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr11_+_119206298 | 0.67 |
ENST00000634586.1
ENST00000634840.1 ENST00000264033.6 ENST00000637974.1 |
CBL
|
Cbl proto-oncogene |
| chr16_+_57092570 | 0.67 |
ENST00000290776.13
ENST00000535318.6 |
CPNE2
|
copine 2 |
| chr20_+_33731976 | 0.67 |
ENST00000375200.6
|
ZNF341
|
zinc finger protein 341 |
| chr10_-_15719885 | 0.66 |
ENST00000378076.4
|
ITGA8
|
integrin subunit alpha 8 |
| chr1_-_172444055 | 0.65 |
ENST00000344529.5
ENST00000367728.1 |
PIGC
|
phosphatidylinositol glycan anchor biosynthesis class C |
| chr11_-_117232033 | 0.65 |
ENST00000524507.6
ENST00000320934.8 ENST00000532301.5 ENST00000676339.1 ENST00000540028.5 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr8_+_22589240 | 0.65 |
ENST00000450780.6
ENST00000430850.6 ENST00000447849.2 ENST00000614502.4 ENST00000443561.3 |
ENSG00000248235.6
PDLIM2
|
novel protein PDZ and LIM domain 2 |
| chr19_-_2051224 | 0.65 |
ENST00000309340.11
ENST00000589534.2 ENST00000250896.9 ENST00000589509.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr8_-_144409282 | 0.64 |
ENST00000620219.4
ENST00000616140.2 |
CPSF1
|
cleavage and polyadenylation specific factor 1 |
| chr6_+_137867241 | 0.64 |
ENST00000612899.5
ENST00000420009.5 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr20_+_31514410 | 0.64 |
ENST00000335574.10
ENST00000340852.9 ENST00000398174.9 ENST00000466766.2 ENST00000498035.5 ENST00000344042.5 |
HM13
|
histocompatibility minor 13 |
| chr1_+_145845608 | 0.63 |
ENST00000334513.6
|
NUDT17
|
nudix hydrolase 17 |
| chr1_-_226408045 | 0.63 |
ENST00000366794.10
ENST00000677203.1 |
PARP1
|
poly(ADP-ribose) polymerase 1 |
| chr18_+_46174014 | 0.63 |
ENST00000619301.4
ENST00000615052.5 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr2_+_90021567 | 0.61 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr5_+_146447304 | 0.61 |
ENST00000296702.9
ENST00000394421.7 ENST00000679501.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr14_+_21846534 | 0.61 |
ENST00000390434.3
|
TRAV8-2
|
T cell receptor alpha variable 8-2 |
| chr14_+_22226711 | 0.60 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr5_-_141619049 | 0.60 |
ENST00000647433.1
ENST00000253811.10 ENST00000389057.9 ENST00000398557.8 |
DIAPH1
|
diaphanous related formin 1 |
| chr17_-_35119801 | 0.60 |
ENST00000592577.5
ENST00000590016.5 ENST00000345365.11 |
RAD51D
|
RAD51 paralog D |
| chr16_+_15643267 | 0.60 |
ENST00000396355.5
|
NDE1
|
nudE neurodevelopment protein 1 |
| chr4_+_74933108 | 0.60 |
ENST00000307428.7
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr4_-_15963165 | 0.59 |
ENST00000259989.7
|
FGFBP2
|
fibroblast growth factor binding protein 2 |
| chr1_-_6554501 | 0.59 |
ENST00000377705.6
|
NOL9
|
nucleolar protein 9 |
| chr1_+_211259932 | 0.59 |
ENST00000367005.8
|
RCOR3
|
REST corepressor 3 |
| chr2_-_89160329 | 0.59 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr6_+_137867414 | 0.59 |
ENST00000237289.8
ENST00000433680.1 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr3_+_58306505 | 0.59 |
ENST00000461393.7
|
HTD2
|
hydroxyacyl-thioester dehydratase type 2 |
| chr6_-_18122677 | 0.59 |
ENST00000340650.6
|
NHLRC1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr12_-_82358380 | 0.59 |
ENST00000256151.8
ENST00000552377.5 |
CCDC59
|
coiled-coil domain containing 59 |
| chr19_-_10335773 | 0.58 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr3_-_184017863 | 0.58 |
ENST00000427120.6
ENST00000334444.11 ENST00000392579.6 ENST00000382494.6 ENST00000265586.10 ENST00000446941.2 |
ABCC5
|
ATP binding cassette subfamily C member 5 |
| chr10_-_75109106 | 0.58 |
ENST00000607487.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr2_-_20651053 | 0.58 |
ENST00000631166.1
ENST00000402541.5 ENST00000446825.1 ENST00000406618.3 ENST00000304031.8 |
HS1BP3
|
HCLS1 binding protein 3 |
| chr14_-_93115812 | 0.57 |
ENST00000553452.5
|
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr17_-_42681840 | 0.57 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10 |
| chr9_-_133149334 | 0.57 |
ENST00000393160.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr1_+_10210562 | 0.57 |
ENST00000377093.9
ENST00000676179.1 |
KIF1B
|
kinesin family member 1B |
| chrX_+_11758159 | 0.57 |
ENST00000361672.6
ENST00000337339.7 ENST00000647869.1 ENST00000312196.10 ENST00000647857.1 ENST00000649130.1 |
MSL3
|
MSL complex subunit 3 |
| chr12_-_56189459 | 0.57 |
ENST00000550164.6
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 |
| chr17_-_29180359 | 0.57 |
ENST00000531253.5
ENST00000628822.2 |
MYO18A
|
myosin XVIIIA |
| chr16_-_31202733 | 0.56 |
ENST00000350605.4
ENST00000247470.10 |
PYCARD
|
PYD and CARD domain containing |
| chr8_+_38901757 | 0.56 |
ENST00000616834.1
|
PLEKHA2
|
pleckstrin homology domain containing A2 |
| chr8_+_42271289 | 0.56 |
ENST00000520810.6
ENST00000520835.7 |
IKBKB
|
inhibitor of nuclear factor kappa B kinase subunit beta |
| chr9_-_120477354 | 0.56 |
ENST00000416449.5
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr10_+_97572771 | 0.55 |
ENST00000370655.6
ENST00000455090.1 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr8_+_38901218 | 0.55 |
ENST00000521746.5
ENST00000616927.4 |
PLEKHA2
|
pleckstrin homology domain containing A2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.7 | 2.8 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.7 | 3.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.6 | 2.4 | GO:0052255 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.6 | 2.9 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.5 | 4.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 1.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.3 | 1.9 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.3 | 1.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.3 | 0.8 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.3 | 1.0 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.2 | 2.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.9 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 0.6 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 | 1.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.2 | 0.6 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.2 | 1.5 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 1.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 1.7 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 0.6 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.2 | 0.8 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.2 | 0.6 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.2 | 0.6 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 | 1.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 1.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 2.6 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.2 | 4.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 2.4 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 | 0.9 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 2.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 5.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.4 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.1 | 0.6 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.9 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 1.6 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.3 | GO:0032258 | CVT pathway(GO:0032258) |
| 0.1 | 1.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.7 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.4 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 1.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.4 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 1.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.7 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.3 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.8 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.6 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 2.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 1.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.3 | GO:0019483 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.2 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 0.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.5 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.2 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) cytosol to ER transport(GO:0046967) |
| 0.1 | 0.4 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 1.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 0.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.2 | GO:0009946 | proximal/distal axis specification(GO:0009946) semicircular canal formation(GO:0060876) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.6 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.7 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 16.0 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 1.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.3 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.4 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.4 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 1.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 0.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.4 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.4 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.2 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.5 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 1.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.6 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.3 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.9 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 8.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.5 | GO:0000050 | urea cycle(GO:0000050) cellular response to lithium ion(GO:0071285) |
| 0.0 | 2.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.5 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:1901838 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 1.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.6 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.5 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 1.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 3.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 2.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 2.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.3 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.6 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 1.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 2.0 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.3 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 1.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.9 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 1.0 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 2.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 1.0 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 1.0 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 1.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 2.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.0 | GO:0060702 | regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 1.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.0 | GO:1904828 | phenotypic switching(GO:0036166) regulation of phenotypic switching by transcription from RNA polymerase II promoter(GO:0100057) regulation of phenotypic switching(GO:1900239) regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.3 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 2.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:2000984 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 1.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 2.7 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 4.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 3.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.4 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 2.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.6 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.7 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 1.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 8.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 2.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.8 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.8 | GO:0090543 | ESCRT III complex(GO:0000815) Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 4.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.0 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.4 | 1.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.4 | 2.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 2.8 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.3 | 4.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 0.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 3.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 2.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 0.7 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.2 | 1.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 2.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.2 | GO:0046977 | peptide antigen-transporting ATPase activity(GO:0015433) TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 0.2 | 4.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 1.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 5.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.5 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 0.7 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.2 | 2.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.5 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.2 | 0.5 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 0.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.4 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.1 | 0.6 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 3.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.0 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.1 | 0.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.6 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 27.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.7 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.1 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.3 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.6 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.2 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 1.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 1.1 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 2.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 2.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 6.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.9 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.4 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 4.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 5.7 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 4.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 3.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 4.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 4.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 4.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 2.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 4.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 9.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.5 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 2.1 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |