Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for RARB
Z-value: 1.65
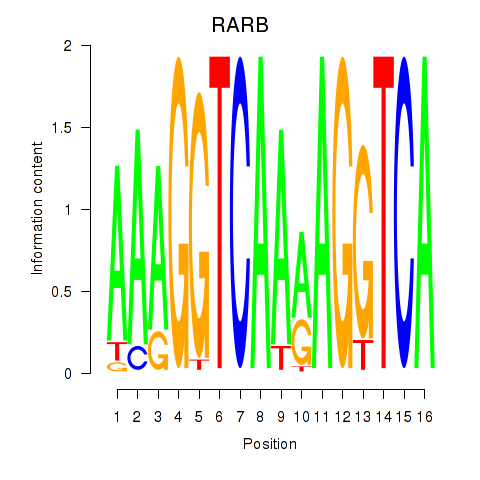
Transcription factors associated with RARB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARB
|
ENSG00000077092.19 | RARB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARB | hg38_v1_chr3_+_25428233_25428284 | -0.03 | 8.7e-01 | Click! |
Activity profile of RARB motif
Sorted Z-values of RARB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RARB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_230745574 | 4.91 |
ENST00000681269.1
|
AGT
|
angiotensinogen |
| chr2_+_17540670 | 4.71 |
ENST00000451533.5
ENST00000295156.9 |
VSNL1
|
visinin like 1 |
| chr2_-_21044063 | 4.29 |
ENST00000233242.5
|
APOB
|
apolipoprotein B |
| chr12_+_57550027 | 3.69 |
ENST00000674619.1
ENST00000676359.1 ENST00000286452.5 ENST00000455537.7 ENST00000676457.1 |
KIF5A
|
kinesin family member 5A |
| chr5_-_135954962 | 3.24 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr4_-_68670648 | 2.98 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr11_-_116837586 | 2.94 |
ENST00000375320.5
ENST00000359492.6 ENST00000375329.6 ENST00000375323.5 ENST00000236850.5 |
APOA1
|
apolipoprotein A1 |
| chr17_-_28576882 | 2.90 |
ENST00000395319.7
ENST00000581807.5 ENST00000226253.9 ENST00000584086.5 ENST00000395321.6 |
ALDOC
|
aldolase, fructose-bisphosphate C |
| chr4_-_6555609 | 2.75 |
ENST00000507294.1
|
PPP2R2C
|
protein phosphatase 2 regulatory subunit Bgamma |
| chr15_-_53759634 | 2.71 |
ENST00000557913.5
ENST00000360509.10 |
WDR72
|
WD repeat domain 72 |
| chr14_+_94612383 | 2.65 |
ENST00000393080.8
ENST00000555820.1 ENST00000393078.5 ENST00000467132.5 |
SERPINA3
|
serpin family A member 3 |
| chr18_-_72543528 | 2.55 |
ENST00000585159.5
ENST00000584764.5 |
CBLN2
|
cerebellin 2 precursor |
| chr16_+_20451563 | 2.40 |
ENST00000417235.6
ENST00000219054.10 |
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A |
| chr6_-_49463173 | 2.34 |
ENST00000274813.4
|
MMUT
|
methylmalonyl-CoA mutase |
| chr6_+_24494939 | 2.33 |
ENST00000348925.2
ENST00000357578.8 |
ALDH5A1
|
aldehyde dehydrogenase 5 family member A1 |
| chr7_-_44141285 | 2.30 |
ENST00000458240.5
ENST00000223364.7 |
MYL7
|
myosin light chain 7 |
| chr14_-_80231052 | 2.20 |
ENST00000557010.5
|
DIO2
|
iodothyronine deiodinase 2 |
| chr11_-_116792386 | 2.18 |
ENST00000433069.2
ENST00000542499.5 |
APOA5
|
apolipoprotein A5 |
| chr16_+_20451596 | 2.07 |
ENST00000575690.5
ENST00000571894.1 |
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A |
| chr4_-_175907143 | 2.04 |
ENST00000513365.1
ENST00000513667.5 ENST00000503563.1 |
GPM6A
|
glycoprotein M6A |
| chr2_-_21043941 | 2.00 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr4_-_170090153 | 1.93 |
ENST00000509167.5
ENST00000353187.6 ENST00000507375.5 ENST00000515480.5 |
AADAT
|
aminoadipate aminotransferase |
| chr12_-_9999176 | 1.92 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr10_-_100065394 | 1.91 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N subunit 1 |
| chr10_-_107164692 | 1.87 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1 |
| chr11_+_73648979 | 1.85 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr11_+_27055215 | 1.84 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr11_+_107591077 | 1.73 |
ENST00000531234.5
ENST00000265840.12 |
ELMOD1
|
ELMO domain containing 1 |
| chr1_+_52927254 | 1.72 |
ENST00000371514.8
ENST00000528311.5 ENST00000371509.8 ENST00000407246.6 ENST00000371513.9 |
SCP2
|
sterol carrier protein 2 |
| chr6_+_24494839 | 1.71 |
ENST00000491546.5
|
ALDH5A1
|
aldehyde dehydrogenase 5 family member A1 |
| chr9_-_101383558 | 1.71 |
ENST00000674556.1
|
BAAT
|
bile acid-CoA:amino acid N-acyltransferase |
| chr7_-_99784248 | 1.71 |
ENST00000652018.1
|
CYP3A4
|
cytochrome P450 family 3 subfamily A member 4 |
| chr10_+_66926028 | 1.70 |
ENST00000361320.5
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr1_+_59310071 | 1.69 |
ENST00000371212.5
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr2_-_151973991 | 1.67 |
ENST00000534999.6
ENST00000637217.1 ENST00000360283.11 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr19_-_50513870 | 1.66 |
ENST00000389208.9
|
ASPDH
|
aspartate dehydrogenase domain containing |
| chr11_-_624924 | 1.65 |
ENST00000358353.8
ENST00000397542.7 ENST00000526077.5 ENST00000534311.1 ENST00000531088.5 |
CDHR5
|
cadherin related family member 5 |
| chr16_-_70678315 | 1.61 |
ENST00000562883.6
|
MTSS2
|
MTSS I-BAR domain containing 2 |
| chr16_+_29679132 | 1.60 |
ENST00000395384.9
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr1_+_196977550 | 1.59 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr4_-_99219230 | 1.59 |
ENST00000394897.5
ENST00000508558.1 ENST00000394899.6 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr22_+_20774092 | 1.57 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr2_-_151973780 | 1.56 |
ENST00000637514.1
ENST00000636350.1 ENST00000434468.2 ENST00000637762.1 ENST00000637779.1 ENST00000637547.1 ENST00000636901.1 ENST00000397327.7 ENST00000636721.1 ENST00000636380.1 ENST00000637284.1 ENST00000636617.1 ENST00000636947.1 ENST00000638091.1 ENST00000636108.1 ENST00000638040.1 ENST00000636773.1 ENST00000637418.1 ENST00000637216.1 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr11_+_73648889 | 1.53 |
ENST00000542389.5
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr9_+_101533963 | 1.53 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20 |
| chr5_-_77087245 | 1.53 |
ENST00000255198.3
|
ZBED3
|
zinc finger BED-type containing 3 |
| chrX_+_11822423 | 1.46 |
ENST00000656302.1
ENST00000640291.2 |
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr19_-_19273247 | 1.43 |
ENST00000389363.5
|
TM6SF2
|
transmembrane 6 superfamily member 2 |
| chr14_+_94581407 | 1.34 |
ENST00000553511.1
ENST00000329597.12 ENST00000554633.5 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin family A member 5 |
| chr16_+_20451532 | 1.33 |
ENST00000576361.5
|
ACSM2A
|
acyl-CoA synthetase medium chain family member 2A |
| chr2_+_131011683 | 1.31 |
ENST00000355771.7
|
ARHGEF4
|
Rho guanine nucleotide exchange factor 4 |
| chr1_-_109509680 | 1.27 |
ENST00000369864.5
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig like domain 1 |
| chr9_+_101533840 | 1.27 |
ENST00000389120.8
ENST00000374819.6 ENST00000479306.5 |
RNF20
|
ring finger protein 20 |
| chr18_-_46757012 | 1.25 |
ENST00000315087.12
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr2_+_190180835 | 1.25 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr6_+_159761991 | 1.22 |
ENST00000367048.5
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr11_+_89710299 | 1.21 |
ENST00000398290.7
|
TRIM77
|
tripartite motif containing 77 |
| chr10_-_60139366 | 1.21 |
ENST00000373815.5
|
ANK3
|
ankyrin 3 |
| chr13_-_48001305 | 1.19 |
ENST00000643023.1
|
SUCLA2
|
succinate-CoA ligase ADP-forming subunit beta |
| chr6_+_49463360 | 1.18 |
ENST00000335783.4
|
CENPQ
|
centromere protein Q |
| chr1_-_111563934 | 1.17 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr1_-_11262530 | 1.12 |
ENST00000361445.9
|
MTOR
|
mechanistic target of rapamycin kinase |
| chr13_-_48001265 | 1.10 |
ENST00000646932.1
ENST00000433022.1 ENST00000470760.2 ENST00000644338.1 ENST00000646602.1 |
SUCLA2
|
succinate-CoA ligase ADP-forming subunit beta |
| chr8_+_132919403 | 1.08 |
ENST00000519178.5
|
TG
|
thyroglobulin |
| chr8_-_140764386 | 1.06 |
ENST00000520151.5
ENST00000519024.5 ENST00000519465.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_+_247965233 | 1.05 |
ENST00000366480.5
|
OR2AK2
|
olfactory receptor family 2 subfamily AK member 2 |
| chr1_-_44674402 | 1.04 |
ENST00000420706.1
ENST00000372235.7 ENST00000372242.7 ENST00000372243.7 ENST00000372244.3 ENST00000372237.8 |
TMEM53
|
transmembrane protein 53 |
| chr8_+_90001448 | 1.04 |
ENST00000519410.5
ENST00000522161.5 ENST00000220764.7 ENST00000517761.5 ENST00000520227.1 |
DECR1
|
2,4-dienoyl-CoA reductase 1 |
| chr5_+_77086682 | 1.04 |
ENST00000643365.1
ENST00000645183.1 ENST00000645374.1 ENST00000647364.1 ENST00000643848.1 ENST00000643603.1 ENST00000645459.1 ENST00000643269.1 ENST00000503969.6 ENST00000646262.1 |
ZBED3-AS1
ENSG00000284762.1
|
ZBED3 antisense RNA 1 phosphodiesterase 8B |
| chr13_-_48001240 | 1.03 |
ENST00000434484.5
|
SUCLA2
|
succinate-CoA ligase ADP-forming subunit beta |
| chr17_+_47941694 | 1.00 |
ENST00000584061.6
|
PNPO
|
pyridoxamine 5'-phosphate oxidase |
| chr18_-_24272179 | 1.00 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr5_-_132777999 | 1.00 |
ENST00000414594.5
|
SEPTIN8
|
septin 8 |
| chr4_+_76256359 | 1.00 |
ENST00000606246.1
|
FAM47E
|
family with sequence similarity 47 member E |
| chr11_+_61752603 | 0.99 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr19_+_35358460 | 0.99 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3 |
| chr17_-_43125300 | 0.97 |
ENST00000497488.1
ENST00000354071.7 ENST00000489037.1 ENST00000470026.5 ENST00000644555.1 ENST00000586385.5 ENST00000591534.5 ENST00000591849.5 |
BRCA1
|
BRCA1 DNA repair associated |
| chr9_+_66900725 | 0.94 |
ENST00000621410.5
ENST00000621015.4 ENST00000616544.4 ENST00000612867.4 ENST00000619435.1 |
ZNF658
|
zinc finger protein 658 |
| chr20_-_45101112 | 0.94 |
ENST00000306117.5
ENST00000537075.3 |
KCNS1
|
potassium voltage-gated channel modifier subfamily S member 1 |
| chr4_-_6200520 | 0.94 |
ENST00000409021.9
ENST00000409371.8 ENST00000282924.9 ENST00000531445.3 |
JAKMIP1
C4orf50
|
janus kinase and microtubule interacting protein 1 chromosome 4 open reading frame 50 |
| chr11_+_118527463 | 0.94 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36 |
| chr5_+_140841183 | 0.90 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr6_+_10694916 | 0.89 |
ENST00000379568.4
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr14_+_103100328 | 0.88 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3 like 4 |
| chr7_-_56092932 | 0.86 |
ENST00000446428.5
ENST00000432123.5 ENST00000297373.7 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr1_-_46176482 | 0.85 |
ENST00000540385.2
ENST00000506599.2 |
P3R3URF-PIK3R3
P3R3URF
|
P3R3URF-PIK3R3 readthrough PIK3R3 upstream reading frame |
| chr1_-_178496939 | 0.83 |
ENST00000623247.2
|
CLEC20A
|
C-type lectin domain containing 20A |
| chr12_+_28452493 | 0.83 |
ENST00000542801.5
|
CCDC91
|
coiled-coil domain containing 91 |
| chr7_-_99735093 | 0.82 |
ENST00000611620.4
ENST00000620220.6 ENST00000336374.4 |
CYP3A7-CYP3A51P
CYP3A7
|
CYP3A7-CYP3A51P readthrough cytochrome P450 family 3 subfamily A member 7 |
| chr11_+_5942251 | 0.82 |
ENST00000641160.1
ENST00000641905.1 |
OR56A3
|
olfactory receptor family 56 subfamily A member 3 |
| chr1_+_10474936 | 0.80 |
ENST00000356607.9
ENST00000491661.2 |
PEX14
|
peroxisomal biogenesis factor 14 |
| chr10_-_27100463 | 0.75 |
ENST00000436985.7
ENST00000376087.5 |
ANKRD26
|
ankyrin repeat domain 26 |
| chr3_-_121729944 | 0.74 |
ENST00000489400.1
|
GOLGB1
|
golgin B1 |
| chr20_+_34874942 | 0.74 |
ENST00000488172.5
|
ACSS2
|
acyl-CoA synthetase short chain family member 2 |
| chr19_-_17448664 | 0.72 |
ENST00000341130.6
|
TMEM221
|
transmembrane protein 221 |
| chr19_-_23003134 | 0.72 |
ENST00000594710.2
|
ZNF728
|
zinc finger protein 728 |
| chr9_+_105700953 | 0.72 |
ENST00000374688.5
|
TMEM38B
|
transmembrane protein 38B |
| chr1_+_16004228 | 0.72 |
ENST00000329454.2
|
SRARP
|
steroid receptor associated and regulated protein |
| chr3_-_64023986 | 0.71 |
ENST00000394431.6
|
PSMD6
|
proteasome 26S subunit, non-ATPase 6 |
| chr5_+_71587351 | 0.71 |
ENST00000683339.1
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 |
| chr6_+_31010474 | 0.71 |
ENST00000561890.1
|
MUC22
|
mucin 22 |
| chr15_+_40239042 | 0.70 |
ENST00000558055.5
ENST00000455577.6 |
PAK6
|
p21 (RAC1) activated kinase 6 |
| chr19_-_22215746 | 0.69 |
ENST00000650058.1
|
ZNF676
|
zinc finger protein 676 |
| chr1_+_86468902 | 0.69 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr16_+_15502266 | 0.68 |
ENST00000452191.6
|
BMERB1
|
bMERB domain containing 1 |
| chr20_-_3204633 | 0.68 |
ENST00000354488.8
ENST00000380201.2 |
DDRGK1
|
DDRGK domain containing 1 |
| chr12_-_119803383 | 0.68 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr3_+_156037693 | 0.67 |
ENST00000472028.5
|
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr7_+_8433602 | 0.66 |
ENST00000405863.6
|
NXPH1
|
neurexophilin 1 |
| chr17_+_80544817 | 0.66 |
ENST00000306801.8
ENST00000570891.5 |
RPTOR
|
regulatory associated protein of MTOR complex 1 |
| chr11_-_89876017 | 0.65 |
ENST00000329862.6
|
TRIM64B
|
tripartite motif containing 64B |
| chr1_-_216805367 | 0.64 |
ENST00000360012.7
|
ESRRG
|
estrogen related receptor gamma |
| chrX_+_153056458 | 0.64 |
ENST00000593810.3
|
PNMA3
|
PNMA family member 3 |
| chr5_+_140794832 | 0.63 |
ENST00000378132.2
ENST00000526136.2 ENST00000520672.2 |
PCDHA2
|
protocadherin alpha 2 |
| chr19_-_32869741 | 0.62 |
ENST00000590341.5
ENST00000587772.1 ENST00000023064.9 |
SLC7A9
|
solute carrier family 7 member 9 |
| chr10_+_121989187 | 0.62 |
ENST00000513429.5
ENST00000515273.5 ENST00000515603.5 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr2_+_190180930 | 0.62 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr13_+_111241185 | 0.62 |
ENST00000478679.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr11_-_49059112 | 0.61 |
ENST00000617704.1
|
TRIM64C
|
tripartite motif containing 64C |
| chr11_+_89710650 | 0.61 |
ENST00000534392.4
|
TRIM77
|
tripartite motif containing 77 |
| chr3_-_47513303 | 0.60 |
ENST00000449409.5
ENST00000414236.5 ENST00000444760.5 ENST00000439305.5 |
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr7_+_99828010 | 0.59 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr1_+_247605586 | 0.58 |
ENST00000320002.3
|
OR2G3
|
olfactory receptor family 2 subfamily G member 3 |
| chr20_-_63969890 | 0.56 |
ENST00000369888.6
|
ZNF512B
|
zinc finger protein 512B |
| chr19_+_35358821 | 0.56 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr10_+_18140412 | 0.55 |
ENST00000324631.13
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr3_+_119597874 | 0.55 |
ENST00000488919.5
ENST00000273371.9 ENST00000495992.5 |
PLA1A
|
phospholipase A1 member A |
| chr8_+_66712700 | 0.55 |
ENST00000521198.7
|
SGK3
|
serum/glucocorticoid regulated kinase family member 3 |
| chr1_-_160343235 | 0.54 |
ENST00000368069.7
ENST00000241704.8 ENST00000647683.1 ENST00000649787.1 |
COPA
|
COPI coat complex subunit alpha |
| chr19_+_13118235 | 0.54 |
ENST00000292431.5
|
NACC1
|
nucleus accumbens associated 1 |
| chr6_+_2987965 | 0.53 |
ENST00000450238.5
ENST00000445000.2 ENST00000426637.5 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr7_+_143052341 | 0.53 |
ENST00000418316.2
|
OR6V1
|
olfactory receptor family 6 subfamily V member 1 |
| chr18_-_77127935 | 0.53 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chrX_-_120575510 | 0.53 |
ENST00000680988.1
ENST00000486604.1 |
CUL4B
|
cullin 4B |
| chr5_-_56116946 | 0.52 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr12_-_103957122 | 0.52 |
ENST00000552940.1
ENST00000547975.5 ENST00000549478.1 ENST00000546540.1 ENST00000378090.9 ENST00000546819.1 ENST00000547945.5 |
C12orf73
|
chromosome 12 open reading frame 73 |
| chr5_-_59216826 | 0.52 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr10_+_121989138 | 0.52 |
ENST00000369005.6
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr17_-_81566582 | 0.51 |
ENST00000572760.5
ENST00000573876.1 |
NPLOC4
|
NPL4 homolog, ubiquitin recognition factor |
| chr1_+_1280436 | 0.51 |
ENST00000379116.10
|
SCNN1D
|
sodium channel epithelial 1 subunit delta |
| chr19_+_18374699 | 0.50 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr22_-_45213492 | 0.50 |
ENST00000424508.5
|
KIAA0930
|
KIAA0930 |
| chr3_-_172711166 | 0.50 |
ENST00000538775.5
ENST00000543711.5 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr19_+_10991175 | 0.49 |
ENST00000644065.1
ENST00000644963.1 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr3_+_63819368 | 0.49 |
ENST00000616659.1
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr1_+_119507203 | 0.47 |
ENST00000369413.8
ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr5_-_77087163 | 0.47 |
ENST00000511587.1
|
ZBED3
|
zinc finger BED-type containing 3 |
| chr3_-_109337572 | 0.46 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4 |
| chr19_+_54803604 | 0.45 |
ENST00000359085.8
|
KIR2DL4
|
killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4 |
| chr17_+_18858068 | 0.45 |
ENST00000536323.5
ENST00000419284.6 ENST00000412418.5 ENST00000575228.5 ENST00000268835.7 ENST00000575102.5 |
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase associated protein 2 |
| chr14_+_20110739 | 0.44 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr17_-_7614824 | 0.44 |
ENST00000571597.1
ENST00000250113.12 |
FXR2
|
FMR1 autosomal homolog 2 |
| chr5_-_97183203 | 0.44 |
ENST00000508447.1
ENST00000283109.8 |
RIOK2
|
RIO kinase 2 |
| chr9_+_72616266 | 0.43 |
ENST00000340019.4
|
TMC1
|
transmembrane channel like 1 |
| chr1_-_202928596 | 0.43 |
ENST00000367258.1
|
KLHL12
|
kelch like family member 12 |
| chr15_+_28919367 | 0.42 |
ENST00000558804.5
|
APBA2
|
amyloid beta precursor protein binding family A member 2 |
| chr1_-_151006795 | 0.42 |
ENST00000312210.9
ENST00000683666.1 |
MINDY1
|
MINDY lysine 48 deubiquitinase 1 |
| chr1_+_1280588 | 0.40 |
ENST00000338555.6
|
SCNN1D
|
sodium channel epithelial 1 subunit delta |
| chr22_-_24593038 | 0.40 |
ENST00000318753.13
|
LRRC75B
|
leucine rich repeat containing 75B |
| chr11_-_111911759 | 0.40 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B |
| chr1_+_119507186 | 0.40 |
ENST00000531340.5
|
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr15_+_90265634 | 0.39 |
ENST00000379095.5
|
NGRN
|
neugrin, neurite outgrowth associated |
| chr19_+_54803535 | 0.39 |
ENST00000396284.6
|
KIR2DL4
|
killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4 |
| chr8_-_23706756 | 0.39 |
ENST00000325017.4
|
NKX2-6
|
NK2 homeobox 6 |
| chr11_+_121576760 | 0.39 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr19_-_15233432 | 0.37 |
ENST00000602233.5
|
EPHX3
|
epoxide hydrolase 3 |
| chr2_+_6978624 | 0.37 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr20_+_54208072 | 0.36 |
ENST00000371419.7
|
PFDN4
|
prefoldin subunit 4 |
| chr2_-_219177137 | 0.36 |
ENST00000453038.5
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr11_-_798261 | 0.36 |
ENST00000530360.2
ENST00000531437.5 ENST00000628067.3 |
SLC25A22
|
solute carrier family 25 member 22 |
| chr16_-_1943259 | 0.36 |
ENST00000622125.4
|
MSRB1
|
methionine sulfoxide reductase B1 |
| chr8_+_13566854 | 0.36 |
ENST00000297324.5
|
C8orf48
|
chromosome 8 open reading frame 48 |
| chr1_-_110407942 | 0.35 |
ENST00000256644.8
|
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr2_-_70553638 | 0.34 |
ENST00000444975.5
ENST00000445399.5 ENST00000295400.11 ENST00000418333.6 |
TGFA
|
transforming growth factor alpha |
| chr14_-_39103187 | 0.34 |
ENST00000548032.6
ENST00000556092.5 ENST00000557280.5 ENST00000307712.11 ENST00000545328.6 ENST00000553970.1 |
SEC23A
|
SEC23 homolog A, COPII coat complex component |
| chr18_+_58196736 | 0.34 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_153150884 | 0.33 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr2_-_55296361 | 0.33 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr2_+_231592858 | 0.33 |
ENST00000313965.4
|
TEX44
|
testis expressed 44 |
| chr11_-_3379212 | 0.33 |
ENST00000429541.6
ENST00000532539.1 ENST00000343338.11 ENST00000620374.4 |
ZNF195
|
zinc finger protein 195 |
| chr3_+_37020333 | 0.33 |
ENST00000616768.5
|
MLH1
|
mutL homolog 1 |
| chrX_-_120575783 | 0.32 |
ENST00000680673.1
|
CUL4B
|
cullin 4B |
| chr14_-_94517844 | 0.32 |
ENST00000341228.2
|
SERPINA12
|
serpin family A member 12 |
| chr16_-_1943123 | 0.31 |
ENST00000473663.1
ENST00000399753.2 ENST00000361871.8 ENST00000564908.1 |
MSRB1
|
methionine sulfoxide reductase B1 |
| chr7_+_107580454 | 0.31 |
ENST00000379117.6
ENST00000473124.1 |
BCAP29
|
B cell receptor associated protein 29 |
| chr18_+_21612274 | 0.31 |
ENST00000579618.1
ENST00000300413.10 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr12_-_112006024 | 0.31 |
ENST00000550800.6
ENST00000550037.5 ENST00000549425.5 |
TMEM116
|
transmembrane protein 116 |
| chr12_-_112005963 | 0.30 |
ENST00000550233.1
|
TMEM116
|
transmembrane protein 116 |
| chr6_+_26500296 | 0.30 |
ENST00000684113.1
|
BTN1A1
|
butyrophilin subfamily 1 member A1 |
| chr3_-_109316505 | 0.29 |
ENST00000478945.1
|
DPPA2
|
developmental pluripotency associated 2 |
| chr1_+_156591741 | 0.29 |
ENST00000368234.7
ENST00000680087.1 ENST00000681734.1 ENST00000679369.1 ENST00000680269.1 ENST00000680661.1 ENST00000681054.1 ENST00000680004.1 ENST00000679702.1 ENST00000368235.8 ENST00000368233.3 |
NAXE
|
NAD(P)HX epimerase |
| chr7_+_107580215 | 0.29 |
ENST00000465919.5
ENST00000005259.9 ENST00000445771.6 ENST00000479917.5 ENST00000421217.5 ENST00000457837.5 |
BCAP29
|
B cell receptor associated protein 29 |
| chrX_-_53434341 | 0.28 |
ENST00000375298.4
ENST00000375304.9 ENST00000684692.1 ENST00000168216.11 |
HSD17B10
|
hydroxysteroid 17-beta dehydrogenase 10 |
| chr11_-_8938211 | 0.28 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr19_+_35370929 | 0.28 |
ENST00000454971.2
|
GPR42
|
G protein-coupled receptor 42 |
| chr11_+_89968502 | 0.28 |
ENST00000533122.3
|
TRIM64
|
tripartite motif containing 64 |
| chr2_-_171066936 | 0.28 |
ENST00000453628.1
ENST00000434911.6 |
TLK1
|
tousled like kinase 1 |
| chr15_+_71810539 | 0.28 |
ENST00000617575.5
ENST00000621098.1 |
NR2E3
|
nuclear receptor subfamily 2 group E member 3 |
| chr2_-_106468326 | 0.27 |
ENST00000304514.11
ENST00000409886.4 |
RGPD3
|
RANBP2 like and GRIP domain containing 3 |
| chr4_+_69280472 | 0.27 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.1 | 2.2 | GO:0010902 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.9 | 3.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.8 | 3.3 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.7 | 9.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.7 | 4.8 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.6 | 1.7 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.5 | 1.6 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.5 | 5.8 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.4 | 1.7 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.4 | 2.8 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.4 | 1.5 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 1.9 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.4 | 1.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.4 | 1.1 | GO:1904204 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.3 | 1.6 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.3 | 1.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.7 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 1.0 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.2 | 0.7 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.2 | 1.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.8 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 1.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 1.0 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 biosynthetic process(GO:0042819) |
| 0.2 | 0.8 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.2 | 2.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.2 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.5 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 2.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.4 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 3.0 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 2.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.7 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.1 | 2.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 2.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 1.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.5 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.3 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.7 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 4.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.2 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.2 | GO:0061580 | colon epithelial cell migration(GO:0061580) intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 1.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.3 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.5 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.9 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 3.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:0060454 | detection of temperature stimulus involved in thermoception(GO:0050960) positive regulation of gastric acid secretion(GO:0060454) response to capsazepine(GO:1901594) |
| 0.0 | 1.9 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 3.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.6 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) regulation of GTP binding(GO:1904424) |
| 0.0 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.9 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.8 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 2.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.3 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 1.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 2.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0060003 | copper ion export(GO:0060003) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.5 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 3.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 1.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.5 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 1.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 3.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.3 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.3 | 2.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 1.6 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 3.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.0 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 1.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.5 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.1 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.7 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.3 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 3.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 2.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 4.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 1.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 17.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 4.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 3.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.6 | 2.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.6 | 1.7 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.6 | 1.7 | GO:0052815 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.5 | 1.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 1.8 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.4 | 1.7 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.4 | 1.7 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.4 | 5.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.3 | 1.0 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.3 | 2.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 1.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.3 | 1.9 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.3 | 1.0 | GO:0004733 | pyridoxamine-phosphate oxidase activity(GO:0004733) |
| 0.2 | 2.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 4.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 1.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.2 | 0.5 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 1.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 0.9 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 3.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 3.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 3.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 4.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.3 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.7 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 2.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 3.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 3.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 5.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 3.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |