Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for RELA
Z-value: 3.50
Transcription factors associated with RELA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELA
|
ENSG00000173039.19 | RELA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELA | hg38_v1_chr11_-_65662931_65662972 | 0.63 | 1.1e-04 | Click! |
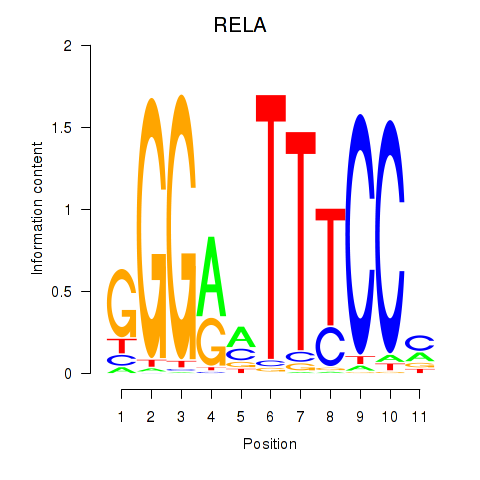
Activity profile of RELA motif
Sorted Z-values of RELA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RELA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 23.9 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 3.2 | 6.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 3.0 | 9.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 2.9 | 8.6 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 2.6 | 10.5 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 2.2 | 8.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.1 | 32.0 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 2.0 | 5.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.9 | 17.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.9 | 1.9 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.8 | 7.0 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.7 | 8.6 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 1.7 | 17.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.7 | 20.4 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 1.6 | 27.6 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 1.6 | 4.8 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.5 | 4.6 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 1.5 | 6.1 | GO:2000753 | regulation of phospholipid scramblase activity(GO:1900161) positive regulation of phospholipid scramblase activity(GO:1900163) regulation of glucosylceramide catabolic process(GO:2000752) positive regulation of glucosylceramide catabolic process(GO:2000753) regulation of sphingomyelin catabolic process(GO:2000754) positive regulation of sphingomyelin catabolic process(GO:2000755) |
| 1.5 | 15.0 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 1.5 | 4.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 1.5 | 5.9 | GO:1990637 | response to prolactin(GO:1990637) |
| 1.3 | 5.2 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 1.2 | 12.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 1.2 | 2.4 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 1.2 | 41.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 1.0 | 3.1 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.0 | 7.9 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.9 | 4.6 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.9 | 4.6 | GO:0097403 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.9 | 8.0 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.9 | 4.3 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.9 | 3.5 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.9 | 4.3 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.8 | 7.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.8 | 4.9 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.8 | 3.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.7 | 4.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.7 | 2.2 | GO:0060264 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.7 | 5.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.7 | 13.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.7 | 2.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.7 | 5.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.7 | 2.0 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.7 | 3.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.6 | 2.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.6 | 7.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.6 | 2.9 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.5 | 2.5 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.5 | 19.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.5 | 2.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.5 | 2.8 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.5 | 7.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.4 | 1.3 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.4 | 6.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 | 0.9 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.4 | 7.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 1.6 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.4 | 2.4 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.4 | 3.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 5.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 1.2 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.4 | 3.9 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.4 | 6.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 4.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.4 | 1.1 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.4 | 3.9 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.4 | 4.6 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.4 | 2.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.3 | 1.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 10.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 12.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.3 | 1.3 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.3 | 1.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 2.4 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.3 | 9.7 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.3 | 27.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 0.9 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.3 | 3.0 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.3 | 3.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 5.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 4.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 6.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.2 | 4.4 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 | 1.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 0.6 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 2.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 5.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.2 | 7.2 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 1.4 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 2.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 23.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.2 | 2.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.2 | 1.7 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 2.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.7 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.2 | 0.3 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.2 | 0.2 | GO:2000152 | negative regulation of protein deubiquitination(GO:0090086) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.2 | 0.8 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 | 3.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 3.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 5.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 2.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 3.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 3.8 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 1.9 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 6.5 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.1 | 1.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 6.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.5 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 3.5 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.1 | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034127) negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) positive regulation of chemokine biosynthetic process(GO:0045080) interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.6 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 4.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.9 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 13.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 3.5 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 3.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 4.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 2.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 1.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 9.2 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 3.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.7 | GO:0060154 | response to cycloheximide(GO:0046898) cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 1.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 4.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 4.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.9 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.9 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 1.5 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.7 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.7 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.9 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 8.1 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 0.2 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 2.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.5 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 5.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 6.8 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.9 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 1.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 4.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.8 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 2.9 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.4 | GO:0034349 | glial cell apoptotic process(GO:0034349) |
| 0.0 | 0.8 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.7 | GO:0032625 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 1.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.0 | 0.7 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 1.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.5 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 | 3.0 | GO:1905037 | autophagosome organization(GO:1905037) |
| 0.0 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 1.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 6.0 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 2.0 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.7 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 3.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.4 | GO:0070671 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 3.6 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.0 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.0 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.2 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 1.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 2.2 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:2000543 | primitive streak formation(GO:0090009) positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.3 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 2.5 | 12.4 | GO:0071754 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 1.8 | 17.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.4 | 4.3 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 1.4 | 15.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.4 | 27.1 | GO:0042611 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 1.3 | 6.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.0 | 6.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.9 | 4.4 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.8 | 5.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.7 | 2.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 2.5 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.5 | 18.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.5 | 4.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 6.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 5.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 2.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 1.3 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.3 | 2.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 1.3 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.3 | 2.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 6.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.3 | 3.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 4.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 0.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 7.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 12.7 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 2.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 11.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 2.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 5.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 59.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 11.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 2.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 8.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 2.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 12.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 4.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 11.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 4.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 4.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 11.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 3.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 5.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.5 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 3.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 18.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 2.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 27.6 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 54.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 4.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 5.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 8.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 25.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.9 | 32.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.6 | 7.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 2.6 | 20.9 | GO:0046979 | TAP2 binding(GO:0046979) |
| 2.3 | 11.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 1.8 | 9.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 1.8 | 7.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 1.7 | 8.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.7 | 8.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.5 | 4.4 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 1.4 | 7.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 1.1 | 18.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 1.1 | 22.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.9 | 4.3 | GO:0004914 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.7 | 4.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.7 | 41.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.7 | 6.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.7 | 4.6 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.6 | 2.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.6 | 5.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.6 | 6.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 26.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.5 | 4.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.5 | 3.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 24.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.5 | 3.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.4 | 4.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.4 | 1.3 | GO:0032428 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.4 | 6.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.4 | 7.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 6.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 1.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.3 | 2.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.3 | 1.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.3 | 2.4 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 2.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 1.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 8.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 6.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.3 | 5.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 10.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 6.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.9 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.2 | 3.0 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 3.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 4.8 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 4.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 3.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 2.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 4.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 2.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 2.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 12.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 2.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.9 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 1.3 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.2 | 9.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 2.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 6.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 6.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 4.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 5.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 6.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 6.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 2.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 1.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 3.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 5.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 4.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 7.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 3.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 7.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 6.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 28.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 2.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 2.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 6.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 1.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 11.5 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 4.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 8.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 4.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 5.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 3.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 3.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 5.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 24.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.1 | 86.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.5 | 8.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 5.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 9.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 7.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.4 | 4.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 36.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.4 | 9.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.3 | 31.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 2.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 23.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 29.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 4.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 11.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 9.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 6.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 13.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 4.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 11.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 6.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 2.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 17.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 34.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 5.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 3.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 2.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 4.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 15.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.0 | 77.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.9 | 55.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.8 | 27.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.7 | 3.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.6 | 14.7 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.6 | 14.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.6 | 14.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.6 | 6.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 24.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.5 | 6.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 9.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 16.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 10.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.3 | 12.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.3 | 14.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 5.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 10.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 5.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 8.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 6.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 7.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 4.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 2.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 4.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 1.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 6.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 5.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 12.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 4.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 7.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 6.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 4.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 5.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 8.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 5.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.2 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 3.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.4 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 1.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |