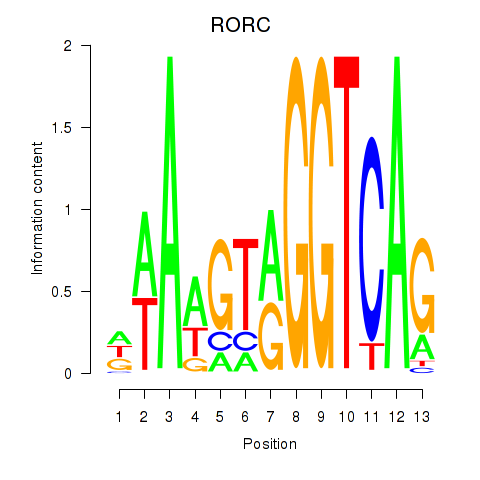
|
chr12_+_7060676
Show fit
|
7.59 |
ENST00000617865.4
ENST00000402681.7
ENST00000360817.10
|
C1S
|
complement C1s
|
|
chr12_+_7061206
Show fit
|
2.66 |
ENST00000423384.5
ENST00000413211.5
|
C1S
|
complement C1s
|
|
chr11_-_64759967
Show fit
|
2.23 |
ENST00000377432.7
ENST00000164139.4
|
PYGM
|
glycogen phosphorylase, muscle associated
|
|
chrX_-_103276741
Show fit
|
2.04 |
ENST00000372680.2
|
TCEAL5
|
transcription elongation factor A like 5
|
|
chr20_-_23452857
Show fit
|
1.68 |
ENST00000377007.3
ENST00000377009.8
|
CST11
|
cystatin 11
|
|
chr19_-_42423100
Show fit
|
1.19 |
ENST00000597001.1
|
LIPE
|
lipase E, hormone sensitive type
|
|
chr11_+_31816266
Show fit
|
1.14 |
ENST00000644607.1
ENST00000646221.1
ENST00000643671.1
ENST00000643931.1
ENST00000642614.1
ENST00000642818.1
ENST00000645848.1
ENST00000506388.2
ENST00000645824.1
ENST00000532942.5
|
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA
novel protein
|
|
chr20_+_38581186
Show fit
|
1.14 |
ENST00000373348.4
ENST00000537425.2
ENST00000416116.2
|
ADIG
|
adipogenin
|
|
chrX_+_70282093
Show fit
|
1.13 |
ENST00000509895.5
ENST00000374473.6
ENST00000276066.4
|
RAB41
|
RAB41, member RAS oncogene family
|
|
chr1_-_235649734
Show fit
|
1.04 |
ENST00000391854.7
|
GNG4
|
G protein subunit gamma 4
|
|
chr19_-_7633713
Show fit
|
0.96 |
ENST00000311069.6
|
PCP2
|
Purkinje cell protein 2
|
|
chr19_-_7632971
Show fit
|
0.94 |
ENST00000598935.5
|
PCP2
|
Purkinje cell protein 2
|
|
chr7_-_141841473
Show fit
|
0.93 |
ENST00000350549.8
ENST00000438520.1
|
PRSS37
|
serine protease 37
|
|
chr6_-_46954922
Show fit
|
0.92 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5
|
|
chr11_-_47594399
Show fit
|
0.82 |
ENST00000302514.4
|
C1QTNF4
|
C1q and TNF related 4
|
|
chr3_-_42410558
Show fit
|
0.77 |
ENST00000441172.1
ENST00000287748.8
|
LYZL4
|
lysozyme like 4
|
|
chr3_-_50322759
Show fit
|
0.72 |
ENST00000442581.1
ENST00000447092.5
|
HYAL2
|
hyaluronidase 2
|
|
chr4_+_168712159
Show fit
|
0.71 |
ENST00000510998.5
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr17_-_10804092
Show fit
|
0.71 |
ENST00000581851.1
|
TMEM238L
|
transmembrane protein 238 like
|
|
chr6_-_119078642
Show fit
|
0.69 |
ENST00000621231.4
ENST00000338891.12
|
FAM184A
|
family with sequence similarity 184 member A
|
|
chr20_+_46894581
Show fit
|
0.69 |
ENST00000357410.7
ENST00000611592.4
|
EYA2
|
EYA transcriptional coactivator and phosphatase 2
|
|
chr3_-_50322733
Show fit
|
0.66 |
ENST00000428028.1
ENST00000357750.9
|
HYAL2
|
hyaluronidase 2
|
|
chr7_-_712940
Show fit
|
0.62 |
ENST00000544935.5
ENST00000430040.5
ENST00000456696.2
ENST00000406797.5
|
PRKAR1B
|
protein kinase cAMP-dependent type I regulatory subunit beta
|
|
chr8_-_94436926
Show fit
|
0.53 |
ENST00000481490.3
|
FSBP
|
fibrinogen silencer binding protein
|
|
chr8_+_39913881
Show fit
|
0.51 |
ENST00000518237.6
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr10_-_104085847
Show fit
|
0.50 |
ENST00000648076.2
|
COL17A1
|
collagen type XVII alpha 1 chain
|
|
chr3_-_65597886
Show fit
|
0.50 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr10_-_101818425
Show fit
|
0.49 |
ENST00000361464.8
ENST00000439817.5
|
OGA
|
O-GlcNAcase
|
|
chr14_+_21317547
Show fit
|
0.47 |
ENST00000557351.1
|
RPGRIP1
|
RPGR interacting protein 1
|
|
chr12_+_93570381
Show fit
|
0.44 |
ENST00000549206.5
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr12_+_93569814
Show fit
|
0.42 |
ENST00000340600.6
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr10_-_101818405
Show fit
|
0.40 |
ENST00000357797.9
ENST00000370094.7
|
OGA
|
O-GlcNAcase
|
|
chr8_+_66043413
Show fit
|
0.39 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta
|
|
chr6_-_136526654
Show fit
|
0.38 |
ENST00000611373.1
|
MAP7
|
microtubule associated protein 7
|
|
chr8_-_126557691
Show fit
|
0.35 |
ENST00000652209.1
|
LRATD2
|
LRAT domain containing 2
|
|
chr14_+_21317535
Show fit
|
0.35 |
ENST00000382933.8
|
RPGRIP1
|
RPGR interacting protein 1
|
|
chr20_-_41317602
Show fit
|
0.34 |
ENST00000559234.5
ENST00000683867.1
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr20_+_31637905
Show fit
|
0.34 |
ENST00000376075.4
|
COX4I2
|
cytochrome c oxidase subunit 4I2
|
|
chr6_-_111605859
Show fit
|
0.34 |
ENST00000651359.1
ENST00000650859.1
ENST00000359831.8
ENST00000368761.11
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_-_157099798
Show fit
|
0.31 |
ENST00000454449.3
|
ETV3L
|
ETS variant transcription factor 3 like
|
|
chr3_-_45915698
Show fit
|
0.28 |
ENST00000539217.5
|
LZTFL1
|
leucine zipper transcription factor like 1
|
|
chr3_+_185586270
Show fit
|
0.28 |
ENST00000296257.10
|
SENP2
|
SUMO specific peptidase 2
|
|
chr1_+_103655760
Show fit
|
0.27 |
ENST00000370083.9
|
AMY1A
|
amylase alpha 1A
|
|
chr19_-_39245006
Show fit
|
0.26 |
ENST00000413851.3
ENST00000613087.4
|
IFNL3
|
interferon lambda 3
|
|
chr4_-_86849366
Show fit
|
0.25 |
ENST00000273905.7
|
SLC10A6
|
solute carrier family 10 member 6
|
|
chr17_-_61863327
Show fit
|
0.25 |
ENST00000584322.2
ENST00000682369.1
ENST00000683039.1
ENST00000683381.1
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr16_+_1782940
Show fit
|
0.24 |
ENST00000262302.14
ENST00000563136.5
ENST00000565987.5
ENST00000568287.5
ENST00000565134.5
|
NUBP2
|
nucleotide binding protein 2
|
|
chr14_-_23286038
Show fit
|
0.24 |
ENST00000673724.1
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr11_+_67605488
Show fit
|
0.23 |
ENST00000533876.1
ENST00000647561.1
|
ENSG00000255119.1
NDUFV1
|
novel transcript
NADH:ubiquinone oxidoreductase core subunit V1
|
|
chr19_-_46077093
Show fit
|
0.23 |
ENST00000601672.5
|
IGFL4
|
IGF like family member 4
|
|
chr5_+_178130996
Show fit
|
0.23 |
ENST00000515098.5
ENST00000502814.5
ENST00000313386.9
ENST00000507457.5
ENST00000508647.5
|
RMND5B
|
required for meiotic nuclear division 5 homolog B
|
|
chr6_+_43770707
Show fit
|
0.22 |
ENST00000324450.11
ENST00000417285.7
ENST00000413642.8
ENST00000372055.9
ENST00000482630.7
ENST00000425836.7
ENST00000372064.9
ENST00000372077.8
ENST00000519767.5
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr19_+_39268394
Show fit
|
0.21 |
ENST00000331982.6
|
IFNL2
|
interferon lambda 2
|
|
chr6_+_43770202
Show fit
|
0.21 |
ENST00000372067.8
ENST00000672860.2
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr13_+_108596152
Show fit
|
0.20 |
ENST00000356711.7
ENST00000251041.10
|
MYO16
|
myosin XVI
|
|
chr2_-_148020689
Show fit
|
0.16 |
ENST00000457954.5
ENST00000392857.10
ENST00000540442.5
ENST00000535373.5
|
ORC4
|
origin recognition complex subunit 4
|
|
chr3_-_165196689
Show fit
|
0.16 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3
|
|
chr1_+_159826799
Show fit
|
0.16 |
ENST00000368104.4
|
SLAMF8
|
SLAM family member 8
|
|
chr3_-_184378136
Show fit
|
0.16 |
ENST00000445696.6
|
THPO
|
thrombopoietin
|
|
chr1_+_154567770
Show fit
|
0.16 |
ENST00000368476.4
|
CHRNB2
|
cholinergic receptor nicotinic beta 2 subunit
|
|
chr19_+_1269266
Show fit
|
0.16 |
ENST00000585630.5
ENST00000589710.5
ENST00000628979.2
ENST00000586773.5
ENST00000587323.5
ENST00000589686.5
ENST00000588230.5
ENST00000413636.6
ENST00000587896.6
ENST00000320936.9
ENST00000589235.5
ENST00000591659.5
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr2_-_29921580
Show fit
|
0.16 |
ENST00000389048.8
|
ALK
|
ALK receptor tyrosine kinase
|
|
chr2_-_148020754
Show fit
|
0.15 |
ENST00000440042.1
ENST00000536575.5
|
ORC4
|
origin recognition complex subunit 4
|
|
chr12_+_56021005
Show fit
|
0.14 |
ENST00000547167.6
|
IKZF4
|
IKAROS family zinc finger 4
|
|
chr14_-_23286082
Show fit
|
0.14 |
ENST00000357460.7
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr1_-_61725121
Show fit
|
0.13 |
ENST00000371177.2
ENST00000606498.5
|
TM2D1
|
TM2 domain containing 1
|
|
chr3_-_165196369
Show fit
|
0.13 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3
|
|
chr14_-_53951880
Show fit
|
0.12 |
ENST00000609748.1
ENST00000558961.1
|
BMP4
|
bone morphogenetic protein 4
|
|
chr22_-_32464440
Show fit
|
0.11 |
ENST00000397450.2
ENST00000397452.5
ENST00000300399.8
|
BPIFC
|
BPI fold containing family C
|
|
chr2_+_148021083
Show fit
|
0.11 |
ENST00000642680.2
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr7_-_128775793
Show fit
|
0.10 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive
|
|
chr1_+_159826860
Show fit
|
0.10 |
ENST00000289707.10
|
SLAMF8
|
SLAM family member 8
|
|
chr12_+_4562192
Show fit
|
0.08 |
ENST00000543431.5
|
DYRK4
|
dual specificity tyrosine phosphorylation regulated kinase 4
|
|
chr3_+_187743654
Show fit
|
0.08 |
ENST00000648485.1
|
ENSG00000285938.1
|
BCL6 antisense 1
|
|
chr10_+_100535927
Show fit
|
0.07 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor
|
|
chr10_-_104085793
Show fit
|
0.04 |
ENST00000650263.1
|
COL17A1
|
collagen type XVII alpha 1 chain
|
|
chr16_-_69351778
Show fit
|
0.03 |
ENST00000288025.4
|
TMED6
|
transmembrane p24 trafficking protein 6
|
|
chr7_-_30550761
Show fit
|
0.01 |
ENST00000598361.4
ENST00000440438.6
|
ENSG00000281039.1
GARS1-DT
|
novel protein
GARS1 divergent transcript
|
|
chr12_+_12908961
Show fit
|
0.01 |
ENST00000540125.1
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A
|