Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
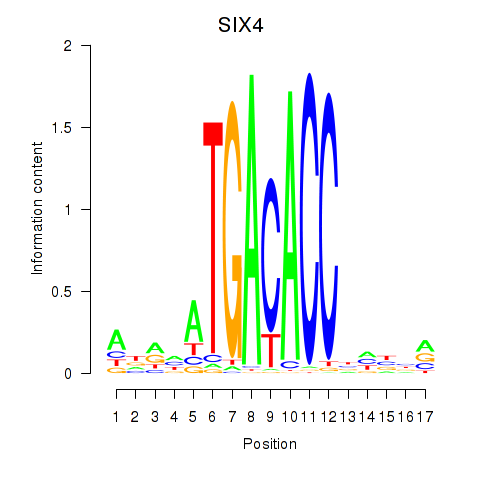
Results for SIX4
Z-value: 0.82
Transcription factors associated with SIX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX4
|
ENSG00000100625.9 | SIX4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX4 | hg38_v1_chr14_-_60724300_60724362 | 0.15 | 4.2e-01 | Click! |
Activity profile of SIX4 motif
Sorted Z-values of SIX4 motif
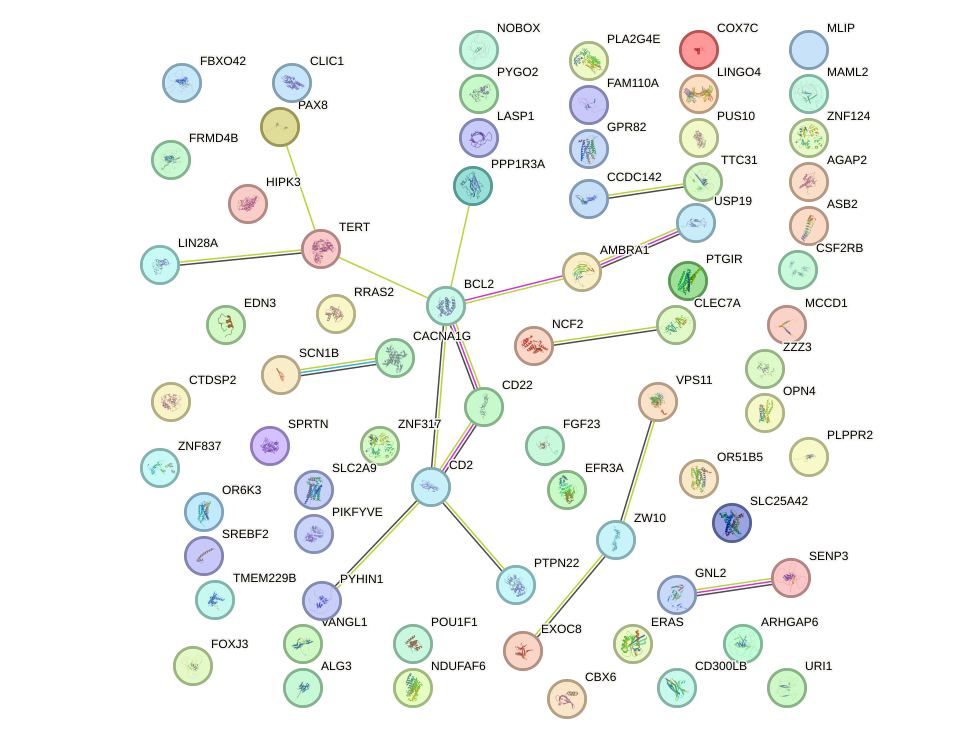
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_116754422 | 2.59 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr14_-_106627685 | 2.57 |
ENST00000390629.3
|
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr14_-_106235582 | 2.27 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr14_-_106324743 | 2.25 |
ENST00000390612.3
|
IGHV4-28
|
immunoglobulin heavy variable 4-28 |
| chr14_-_106349792 | 2.18 |
ENST00000438142.3
|
IGHV4-31
|
immunoglobulin heavy variable 4-31 |
| chr14_-_105940235 | 1.93 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr14_-_106803221 | 1.55 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chrY_+_20575792 | 1.50 |
ENST00000382772.3
|
EIF1AY
|
eukaryotic translation initiation factor 1A Y-linked |
| chrY_+_20575716 | 1.48 |
ENST00000361365.7
|
EIF1AY
|
eukaryotic translation initiation factor 1A Y-linked |
| chr19_+_35329161 | 1.46 |
ENST00000341773.10
ENST00000600131.5 ENST00000595780.5 ENST00000597916.5 ENST00000593867.5 ENST00000600424.5 ENST00000599811.5 ENST00000536635.6 ENST00000085219.10 ENST00000544992.6 ENST00000419549.6 |
CD22
|
CD22 molecule |
| chr1_+_158930778 | 1.19 |
ENST00000458222.5
|
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr15_-_21742799 | 1.09 |
ENST00000622410.2
|
ENSG00000278263.2
|
novel protein, identical to IGHV4-4 |
| chr12_-_10130143 | 1.04 |
ENST00000298523.9
ENST00000396484.6 ENST00000310002.4 ENST00000304084.13 |
CLEC7A
|
C-type lectin domain containing 7A |
| chrX_+_41724174 | 1.03 |
ENST00000302548.5
|
GPR82
|
G protein-coupled receptor 82 |
| chr10_+_10798570 | 1.01 |
ENST00000638035.1
ENST00000636488.1 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr12_-_10130241 | 0.99 |
ENST00000353231.9
ENST00000525605.1 |
CLEC7A
|
C-type lectin domain containing 7A |
| chr17_-_74531467 | 0.99 |
ENST00000314401.3
ENST00000392621.6 |
CD300LB
|
CD300 molecule like family member b |
| chr19_+_35031263 | 0.94 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr14_+_21924033 | 0.93 |
ENST00000390440.2
|
TRAV14DV4
|
T cell receptor alpha variable 14/delta variable 4 |
| chr15_+_40953463 | 0.89 |
ENST00000617768.5
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chr1_-_151805419 | 0.86 |
ENST00000368820.4
|
LINGO4
|
leucine rich repeat and Ig domain containing 4 |
| chr20_+_836052 | 0.85 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110 member A |
| chr7_-_113919000 | 0.84 |
ENST00000284601.4
|
PPP1R3A
|
protein phosphatase 1 regulatory subunit 3A |
| chr22_+_36922040 | 0.82 |
ENST00000406230.5
|
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr12_-_57846686 | 0.78 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr1_-_113871665 | 0.78 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr19_+_19063986 | 0.77 |
ENST00000318596.8
|
SLC25A42
|
solute carrier family 25 member 42 |
| chr19_-_46625037 | 0.68 |
ENST00000596260.1
ENST00000594275.1 ENST00000597185.1 ENST00000598865.5 ENST00000291294.7 |
PTGIR
|
prostaglandin I2 receptor |
| chr12_-_4379712 | 0.67 |
ENST00000237837.2
|
FGF23
|
fibroblast growth factor 23 |
| chr12_-_57742120 | 0.61 |
ENST00000257897.7
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr12_-_10130082 | 0.59 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr11_-_130314686 | 0.54 |
ENST00000525842.5
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr6_+_53929982 | 0.53 |
ENST00000505762.1
ENST00000511369.5 ENST00000431554.2 |
MLIP
ENSG00000236740.6
|
muscular LMNA interacting protein novel transcript |
| chr11_-_130314575 | 0.52 |
ENST00000397753.5
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr15_+_40952962 | 0.51 |
ENST00000444189.7
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chr1_-_231337830 | 0.50 |
ENST00000366645.1
|
EXOC8
|
exocyst complex component 8 |
| chr1_-_154961720 | 0.44 |
ENST00000368457.3
|
PYGO2
|
pygopus family PHD finger 2 |
| chr11_+_33258304 | 0.42 |
ENST00000531504.5
ENST00000456517.2 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr18_-_63161848 | 0.42 |
ENST00000590515.1
|
BCL2
|
BCL2 apoptosis regulator |
| chr1_-_16352420 | 0.40 |
ENST00000375592.8
|
FBXO42
|
F-box protein 42 |
| chr19_+_50384323 | 0.40 |
ENST00000599857.7
ENST00000613923.6 ENST00000601098.6 ENST00000440232.7 ENST00000595904.6 ENST00000593887.1 |
POLD1
|
DNA polymerase delta 1, catalytic subunit |
| chr1_-_183590876 | 0.39 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr14_-_93955577 | 0.37 |
ENST00000555507.5
|
ASB2
|
ankyrin repeat and SOCS box containing 2 |
| chr19_+_9140377 | 0.37 |
ENST00000360385.7
ENST00000247956.11 |
ZNF317
|
zinc finger protein 317 |
| chr14_-_67515153 | 0.35 |
ENST00000555994.6
|
TMEM229B
|
transmembrane protein 229B |
| chr1_+_26410809 | 0.34 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr20_+_59300589 | 0.34 |
ENST00000337938.7
ENST00000371025.7 |
EDN3
|
endothelin 3 |
| chr8_+_131904071 | 0.32 |
ENST00000254624.10
ENST00000522709.5 |
EFR3A
|
EFR3 homolog A |
| chr1_-_42335869 | 0.32 |
ENST00000372573.5
|
FOXJ3
|
forkhead box J3 |
| chr17_+_50561010 | 0.31 |
ENST00000360761.8
ENST00000354983.8 ENST00000352832.9 |
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chr8_+_94895763 | 0.31 |
ENST00000523378.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr1_+_231338242 | 0.30 |
ENST00000008440.9
ENST00000295050.12 |
SPRTN
|
SprT-like N-terminal domain |
| chr17_+_38870050 | 0.30 |
ENST00000318008.11
ENST00000435347.7 |
LASP1
|
LIM and SH3 protein 1 |
| chr3_-_87276462 | 0.28 |
ENST00000561167.5
ENST00000560656.1 |
POU1F1
|
POU class 1 homeobox 1 |
| chr3_-_69200711 | 0.28 |
ENST00000478263.5
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr3_-_49120785 | 0.27 |
ENST00000417901.5
ENST00000306026.5 ENST00000434032.6 |
USP19
|
ubiquitin specific peptidase 19 |
| chr5_+_86617919 | 0.27 |
ENST00000247655.4
ENST00000509578.1 |
COX7C
|
cytochrome c oxidase subunit 7C |
| chr3_-_49120668 | 0.26 |
ENST00000465902.1
|
USP19
|
ubiquitin specific peptidase 19 |
| chr20_+_59300547 | 0.26 |
ENST00000644821.1
|
EDN3
|
endothelin 3 |
| chr15_+_58987652 | 0.24 |
ENST00000348370.9
ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr3_-_87276577 | 0.24 |
ENST00000344265.8
ENST00000350375.7 |
POU1F1
|
POU class 1 homeobox 1 |
| chr2_-_61017174 | 0.23 |
ENST00000407787.5
ENST00000398658.2 |
PUS10
|
pseudouridine synthase 10 |
| chr2_-_74482914 | 0.23 |
ENST00000290418.4
ENST00000393965.8 |
CCDC142
|
coiled-coil domain containing 142 |
| chr20_+_59300402 | 0.23 |
ENST00000311585.11
ENST00000371028.6 |
EDN3
|
endothelin 3 |
| chr1_-_247172002 | 0.21 |
ENST00000491356.5
ENST00000472531.5 ENST00000340684.10 ENST00000543802.3 |
ZNF124
|
zinc finger protein 124 |
| chr7_-_144403693 | 0.19 |
ENST00000645489.1
ENST00000643164.1 |
NOBOX
|
NOBOX oogenesis homeobox |
| chr3_-_184248935 | 0.19 |
ENST00000446569.1
ENST00000397676.8 |
ALG3
|
ALG3 alpha-1,3- mannosyltransferase |
| chr3_-_184249520 | 0.19 |
ENST00000455059.5
ENST00000445626.6 |
ALG3
|
ALG3 alpha-1,3- mannosyltransferase |
| chr2_-_113241683 | 0.18 |
ENST00000468980.3
|
PAX8
|
paired box 8 |
| chr10_+_86654581 | 0.18 |
ENST00000443292.1
|
OPN4
|
opsin 4 |
| chr1_-_42335189 | 0.18 |
ENST00000361776.5
ENST00000445886.5 ENST00000361346.6 |
FOXJ3
|
forkhead box J3 |
| chr17_+_7561899 | 0.18 |
ENST00000321337.12
|
SENP3
|
SUMO specific peptidase 3 |
| chr19_+_12687998 | 0.17 |
ENST00000640151.1
ENST00000640117.1 |
GNG14
|
G protein subunit gamma 14 |
| chr8_+_94895837 | 0.17 |
ENST00000519136.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr15_-_42051190 | 0.17 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2 group IVE |
| chr6_+_31528956 | 0.17 |
ENST00000376191.3
|
MCCD1
|
mitochondrial coiled-coil domain 1 |
| chr6_-_27872334 | 0.16 |
ENST00000616365.2
|
H3C11
|
H3 clustered histone 11 |
| chr11_-_46593948 | 0.15 |
ENST00000533727.5
ENST00000534300.5 ENST00000683756.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy and beclin 1 regulator 1 |
| chr19_+_29923644 | 0.15 |
ENST00000360605.8
ENST00000570564.5 ENST00000574233.5 ENST00000585655.1 |
URI1
|
URI1 prefoldin like chaperone |
| chr2_+_74483067 | 0.14 |
ENST00000233623.11
ENST00000442235.6 ENST00000410003.5 |
TTC31
|
tetratricopeptide repeat domain 31 |
| chr1_-_37595927 | 0.13 |
ENST00000373062.8
|
GNL2
|
G protein nucleolar 2 |
| chr19_-_58381022 | 0.12 |
ENST00000427624.2
ENST00000597582.2 |
ZNF837
|
zinc finger protein 837 |
| chr11_+_119067774 | 0.11 |
ENST00000621676.5
ENST00000614944.4 |
VPS11
|
VPS11 core subunit of CORVET and HOPS complexes |
| chr22_+_41833079 | 0.11 |
ENST00000612482.4
ENST00000361204.9 |
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr5_-_1294989 | 0.11 |
ENST00000334602.10
ENST00000310581.10 |
TERT
|
telomerase reverse transcriptase |
| chr19_+_859654 | 0.11 |
ENST00000592860.2
ENST00000327726.11 |
CFD
|
complement factor D |
| chr10_+_86654541 | 0.10 |
ENST00000241891.10
ENST00000372071.6 |
OPN4
|
opsin 4 |
| chr1_-_158720720 | 0.10 |
ENST00000368145.2
|
OR6K3
|
olfactory receptor family 6 subfamily K member 3 |
| chr11_-_5343524 | 0.09 |
ENST00000300773.3
|
OR51B5
|
olfactory receptor family 51 subfamily B member 5 |
| chr16_+_590200 | 0.09 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr1_-_224437028 | 0.09 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr5_+_86617967 | 0.08 |
ENST00000515763.1
|
COX7C
|
cytochrome c oxidase subunit 7C |
| chr6_+_52186373 | 0.08 |
ENST00000648244.1
|
IL17A
|
interleukin 17A |
| chrX_+_48828873 | 0.07 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr22_-_38872206 | 0.07 |
ENST00000407418.8
ENST00000216083.6 |
CBX6
|
chromobox 6 |
| chr4_-_10021490 | 0.07 |
ENST00000264784.8
|
SLC2A9
|
solute carrier family 2 member 9 |
| chr11_-_14359118 | 0.07 |
ENST00000256196.9
|
RRAS2
|
RAS related 2 |
| chrX_+_9560465 | 0.07 |
ENST00000647060.1
|
TBL1X
|
transducin beta like 1 X-linked |
| chr1_-_77682639 | 0.04 |
ENST00000370801.8
ENST00000433749.5 |
ZZZ3
|
zinc finger ZZ-type containing 3 |
| chr11_-_96343170 | 0.04 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr6_-_31736504 | 0.03 |
ENST00000616760.1
ENST00000375784.7 ENST00000375779.6 |
CLIC1
|
chloride intracellular channel 1 |
| chr7_-_111788958 | 0.02 |
ENST00000417165.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr11_-_113773668 | 0.02 |
ENST00000200135.8
|
ZW10
|
zw10 kinetochore protein |
| chrX_-_11665908 | 0.02 |
ENST00000337414.9
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr1_+_115641945 | 0.02 |
ENST00000355485.7
ENST00000369510.8 |
VANGL1
|
VANGL planar cell polarity protein 1 |
| chr19_+_11355386 | 0.00 |
ENST00000251473.9
ENST00000591329.5 ENST00000586380.5 |
PLPPR2
|
phospholipid phosphatase related 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.2 | 0.8 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 0.9 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.7 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.5 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 10.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.5 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 0.4 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.8 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) coenzyme transport(GO:0051182) |
| 0.1 | 1.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.3 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 2.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0030526 | granulocyte macrophage colony-stimulating factor receptor complex(GO:0030526) |
| 0.1 | 8.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 2.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 0.9 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.8 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 8.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.6 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 3.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 4.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 2.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |