Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for SOX21
Z-value: 1.21
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.6 | SOX21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg38_v1_chr13_-_94712505_94712553 | -0.41 | 1.9e-02 | Click! |
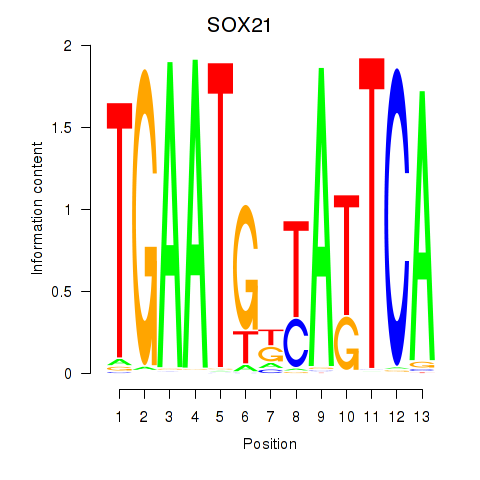
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_41877267 | 5.32 |
ENST00000221972.8
ENST00000597454.1 ENST00000444740.2 |
CD79A
|
CD79a molecule |
| chr11_+_35180279 | 3.92 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_158931539 | 3.91 |
ENST00000368140.6
ENST00000368138.7 ENST00000392254.6 ENST00000392252.7 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family member 1 |
| chr14_+_21941122 | 3.48 |
ENST00000390441.2
|
TRAV9-2
|
T cell receptor alpha variable 9-2 |
| chr22_+_22720615 | 3.45 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr22_+_38957522 | 3.19 |
ENST00000618553.1
ENST00000249116.7 |
APOBEC3A
|
apolipoprotein B mRNA editing enzyme catalytic subunit 3A |
| chr5_-_56233287 | 2.61 |
ENST00000513241.2
ENST00000341048.9 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr17_-_44376169 | 2.28 |
ENST00000587295.5
|
ITGA2B
|
integrin subunit alpha 2b |
| chr7_+_142482544 | 1.97 |
ENST00000390372.3
|
TRBV5-5
|
T cell receptor beta variable 5-5 |
| chr12_-_10435940 | 1.92 |
ENST00000381901.5
ENST00000381902.7 ENST00000539033.1 |
KLRC2
ENSG00000255641.1
|
killer cell lectin like receptor C2 novel protein |
| chr3_-_71306012 | 1.84 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr6_+_131573219 | 1.77 |
ENST00000356962.2
ENST00000368087.8 ENST00000673427.1 ENST00000640973.1 |
ARG1
|
arginase 1 |
| chr11_+_35180342 | 1.73 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr14_+_22226711 | 1.71 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr18_-_49492305 | 1.70 |
ENST00000615479.4
ENST00000583637.5 ENST00000618613.5 ENST00000615760.4 ENST00000578528.1 ENST00000578532.5 ENST00000580387.5 ENST00000579248.5 ENST00000580261.6 ENST00000581373.5 ENST00000618619.4 ENST00000617346.4 ENST00000583036.5 ENST00000332968.11 |
RPL17
RPL17-C18orf32
|
ribosomal protein L17 RPL17-C18orf32 readthrough |
| chr3_-_71305986 | 1.63 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr2_+_216498831 | 1.63 |
ENST00000491306.6
ENST00000600880.5 ENST00000446558.5 |
RPL37A
|
ribosomal protein L37a |
| chr1_+_144592868 | 1.60 |
ENST00000581138.3
|
PPIAL4F
|
peptidylprolyl isomerase A like 4F |
| chr7_+_142535763 | 1.54 |
ENST00000614171.1
|
TRBV13
|
T cell receptor beta variable 13 |
| chr14_-_106470788 | 1.54 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr12_-_89352395 | 1.53 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_89352487 | 1.52 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr6_+_116399395 | 1.51 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr1_+_2556361 | 1.42 |
ENST00000355716.5
|
TNFRSF14
|
TNF receptor superfamily member 14 |
| chr1_+_2556222 | 1.36 |
ENST00000409119.5
|
TNFRSF14
|
TNF receptor superfamily member 14 |
| chr19_+_16197900 | 1.35 |
ENST00000429941.6
ENST00000291439.8 ENST00000444449.6 ENST00000589822.5 |
AP1M1
|
adaptor related protein complex 1 subunit mu 1 |
| chr4_-_39032922 | 1.33 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr7_-_87220520 | 1.30 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243 |
| chr15_-_79971164 | 1.23 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr1_+_149583865 | 1.22 |
ENST00000585245.2
|
PPIAL4C
|
peptidylprolyl isomerase A like 4C |
| chr7_-_87220567 | 1.19 |
ENST00000433078.5
|
TMEM243
|
transmembrane protein 243 |
| chr19_+_16197848 | 1.16 |
ENST00000590263.5
ENST00000590756.5 |
AP1M1
|
adaptor related protein complex 1 subunit mu 1 |
| chr21_+_29300111 | 1.14 |
ENST00000451655.5
|
BACH1
|
BTB domain and CNC homolog 1 |
| chr19_+_18571730 | 1.09 |
ENST00000596304.5
ENST00000430157.6 ENST00000596273.5 ENST00000442744.7 ENST00000595683.5 ENST00000599256.5 ENST00000595158.5 ENST00000598780.5 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr2_+_89851723 | 1.09 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr6_+_42563981 | 1.05 |
ENST00000372899.6
ENST00000372901.2 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr7_+_120989030 | 0.98 |
ENST00000428526.5
|
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr12_-_7109176 | 0.94 |
ENST00000545280.5
ENST00000543933.5 ENST00000545337.1 ENST00000266542.9 ENST00000544702.5 |
C1RL
|
complement C1r subcomponent like |
| chr14_-_92121902 | 0.87 |
ENST00000329559.8
|
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr14_-_92121669 | 0.84 |
ENST00000605997.6
ENST00000553514.5 ENST00000617122.1 |
NDUFB1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chrX_-_132219439 | 0.81 |
ENST00000370874.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr4_-_102825854 | 0.79 |
ENST00000350435.11
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr3_+_155024118 | 0.77 |
ENST00000492661.5
|
MME
|
membrane metalloendopeptidase |
| chr1_+_207325629 | 0.73 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr19_+_44613558 | 0.72 |
ENST00000402988.6
|
IGSF23
|
immunoglobulin superfamily member 23 |
| chr6_+_31403553 | 0.72 |
ENST00000449934.7
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chr3_-_45915596 | 0.71 |
ENST00000472635.5
ENST00000492333.5 |
LZTFL1
|
leucine zipper transcription factor like 1 |
| chr7_+_138076475 | 0.65 |
ENST00000438242.1
|
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr8_-_141367276 | 0.65 |
ENST00000377741.4
|
GPR20
|
G protein-coupled receptor 20 |
| chr6_-_53061740 | 0.60 |
ENST00000350082.10
ENST00000356971.3 ENST00000676107.1 |
CILK1
|
ciliogenesis associated kinase 1 |
| chr11_+_5389377 | 0.59 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr18_+_32190033 | 0.58 |
ENST00000269202.11
|
MEP1B
|
meprin A subunit beta |
| chr1_+_152698339 | 0.57 |
ENST00000368779.2
|
LCE2A
|
late cornified envelope 2A |
| chr12_+_34022462 | 0.57 |
ENST00000538927.1
ENST00000266483.7 |
ALG10
|
ALG10 alpha-1,2-glucosyltransferase |
| chr8_-_65838730 | 0.57 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr7_+_7156934 | 0.56 |
ENST00000429911.5
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1 |
| chr4_-_102825526 | 0.54 |
ENST00000504211.5
ENST00000508476.5 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr18_+_32190015 | 0.50 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chr6_-_154246850 | 0.50 |
ENST00000517438.5
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr3_-_186570308 | 0.49 |
ENST00000446782.5
|
TBCCD1
|
TBCC domain containing 1 |
| chr15_+_58410543 | 0.47 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr3_+_100635598 | 0.46 |
ENST00000475887.1
|
ADGRG7
|
adhesion G protein-coupled receptor G7 |
| chr17_+_68259164 | 0.45 |
ENST00000448504.6
|
ARSG
|
arylsulfatase G |
| chr3_+_186570663 | 0.45 |
ENST00000265028.8
|
DNAJB11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr12_+_55492378 | 0.42 |
ENST00000548615.1
|
OR6C68
|
olfactory receptor family 6 subfamily C member 68 |
| chr12_-_949683 | 0.40 |
ENST00000358495.8
|
RAD52
|
RAD52 homolog, DNA repair protein |
| chr14_+_92121953 | 0.40 |
ENST00000298875.9
ENST00000553427.5 |
CPSF2
|
cleavage and polyadenylation specific factor 2 |
| chr22_-_40533808 | 0.40 |
ENST00000422851.1
ENST00000651694.1 ENST00000652095.2 |
MRTFA
|
myocardin related transcription factor A |
| chr1_-_152089062 | 0.39 |
ENST00000368806.2
|
TCHHL1
|
trichohyalin like 1 |
| chr4_+_68447453 | 0.38 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr7_+_138076453 | 0.37 |
ENST00000242375.8
ENST00000411726.6 |
AKR1D1
|
aldo-keto reductase family 1 member D1 |
| chr1_+_147242654 | 0.36 |
ENST00000652587.1
ENST00000361293.10 ENST00000369258.8 ENST00000369259.4 ENST00000650714.1 ENST00000639534.1 |
CHD1L
|
chromodomain helicase DNA binding protein 1 like |
| chr1_+_200027702 | 0.35 |
ENST00000367362.8
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chrX_+_131083706 | 0.35 |
ENST00000370921.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr1_+_152908538 | 0.34 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr12_-_949519 | 0.33 |
ENST00000397230.6
ENST00000542785.5 ENST00000544742.5 ENST00000536177.5 ENST00000541619.1 |
RAD52
|
RAD52 homolog, DNA repair protein |
| chr1_-_146344790 | 0.33 |
ENST00000584068.3
|
PPIAL4H
|
peptidylprolyl isomerase A like 4H |
| chr1_+_200027605 | 0.31 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2 |
| chr3_+_141384790 | 0.31 |
ENST00000507722.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_-_31760973 | 0.31 |
ENST00000436464.1
|
ADGRB2
|
adhesion G protein-coupled receptor B2 |
| chr4_-_102825767 | 0.28 |
ENST00000505207.5
ENST00000502404.5 ENST00000507845.5 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr17_-_67239800 | 0.27 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr4_-_88159023 | 0.25 |
ENST00000505480.6
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr16_-_18433412 | 0.23 |
ENST00000620440.4
|
NOMO2
|
NODAL modulator 2 |
| chrX_+_11760035 | 0.21 |
ENST00000482871.6
ENST00000648013.1 |
MSL3
|
MSL complex subunit 3 |
| chr1_-_37514743 | 0.21 |
ENST00000448519.2
ENST00000373075.6 ENST00000373073.8 ENST00000296214.10 |
MEAF6
|
MYST/Esa1 associated factor 6 |
| chr12_-_10807286 | 0.21 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr7_+_34658361 | 0.21 |
ENST00000381539.3
|
NPSR1
|
neuropeptide S receptor 1 |
| chr19_-_7040179 | 0.21 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4 |
| chr10_-_17201580 | 0.21 |
ENST00000525762.5
ENST00000488990.5 ENST00000377799.8 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr8_-_142879820 | 0.21 |
ENST00000377675.3
ENST00000517471.5 ENST00000292427.10 |
CYP11B1
|
cytochrome P450 family 11 subfamily B member 1 |
| chr1_+_67166448 | 0.20 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr8_-_86069662 | 0.20 |
ENST00000276616.3
|
PSKH2
|
protein serine kinase H2 |
| chr15_-_34964442 | 0.19 |
ENST00000560117.1
|
AQR
|
aquarius intron-binding spliceosomal factor |
| chrX_-_153944655 | 0.18 |
ENST00000369997.7
|
RENBP
|
renin binding protein |
| chr5_+_176448363 | 0.18 |
ENST00000261942.7
|
FAF2
|
Fas associated factor family member 2 |
| chr8_+_66775178 | 0.18 |
ENST00000396596.2
ENST00000521960.5 ENST00000522398.5 ENST00000522629.5 ENST00000520976.5 |
SGK3
|
serum/glucocorticoid regulated kinase family member 3 |
| chr11_+_60088251 | 0.17 |
ENST00000524868.1
|
MS4A2
|
membrane spanning 4-domains A2 |
| chr1_+_248361665 | 0.15 |
ENST00000366473.4
|
OR2T4
|
olfactory receptor family 2 subfamily T member 4 |
| chr1_+_203795614 | 0.13 |
ENST00000367210.3
ENST00000432282.5 ENST00000453771.5 ENST00000367214.5 ENST00000639812.1 ENST00000367212.7 ENST00000332127.8 ENST00000550078.2 |
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A zinc finger BED-type containing 6 |
| chr5_-_179618032 | 0.13 |
ENST00000519033.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr3_+_98463201 | 0.12 |
ENST00000642057.1
|
OR5K1
|
olfactory receptor family 5 subfamily K member 1 |
| chr7_+_2354810 | 0.12 |
ENST00000360876.9
ENST00000413917.5 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3 subunit B |
| chr8_-_93017656 | 0.10 |
ENST00000520686.5
|
TRIQK
|
triple QxxK/R motif containing |
| chr5_-_60372714 | 0.10 |
ENST00000514552.5
|
PDE4D
|
phosphodiesterase 4D |
| chr11_-_8263858 | 0.10 |
ENST00000534484.1
ENST00000335790.8 |
LMO1
|
LIM domain only 1 |
| chr10_-_13234368 | 0.09 |
ENST00000378681.8
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
| chrX_-_153944621 | 0.09 |
ENST00000393700.8
|
RENBP
|
renin binding protein |
| chr3_-_36992802 | 0.08 |
ENST00000623924.1
|
EPM2AIP1
|
EPM2A interacting protein 1 |
| chr4_-_103719980 | 0.06 |
ENST00000304883.3
|
TACR3
|
tachykinin receptor 3 |
| chr9_+_96928310 | 0.05 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr12_+_55314343 | 0.03 |
ENST00000642104.1
|
OR6C1
|
olfactory receptor family 6 subfamily C member 1 |
| chr6_+_131637296 | 0.03 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr6_+_29396555 | 0.02 |
ENST00000623183.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2 |
| chr10_-_13234329 | 0.01 |
ENST00000463405.2
|
UCMA
|
upper zone of growth plate and cartilage matrix associated |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.4 | 1.8 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.4 | 5.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.4 | 3.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.3 | 2.8 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 1.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 1.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.2 | 3.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.2 | 0.6 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.7 | GO:0035739 | CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.7 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 2.5 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.3 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.7 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 5.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.3 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 7.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 2.3 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 1.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 3.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.9 | GO:0002228 | natural killer cell mediated immunity(GO:0002228) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 2.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 5.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.2 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 4.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.5 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.5 | 2.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 1.9 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.2 | 1.0 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.2 | 0.6 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 3.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 5.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.2 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 2.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 6.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 4.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 6.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 5.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 2.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 3.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |