Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
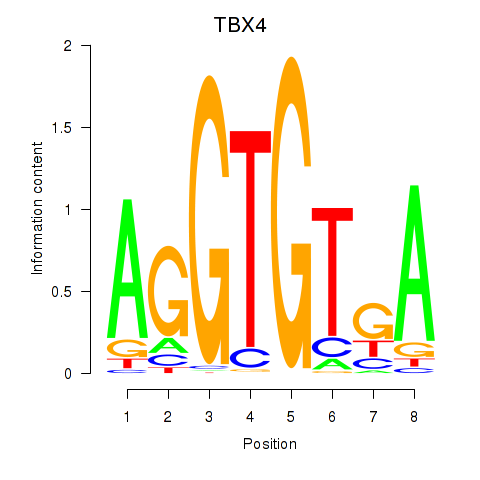
Results for TBX4
Z-value: 1.24
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.11 | TBX4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX4 | hg38_v1_chr17_+_61452378_61452437 | -0.28 | 1.2e-01 | Click! |
Activity profile of TBX4 motif
Sorted Z-values of TBX4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_115180037 | 2.39 |
ENST00000514154.1
ENST00000282369.7 |
TRIM36
|
tripartite motif containing 36 |
| chr12_+_18738102 | 1.93 |
ENST00000317658.5
|
CAPZA3
|
capping actin protein of muscle Z-line subunit alpha 3 |
| chr8_+_102551583 | 1.86 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr10_+_44911316 | 1.85 |
ENST00000544540.5
ENST00000389583.5 |
TMEM72
|
transmembrane protein 72 |
| chr6_-_130215629 | 1.69 |
ENST00000532763.5
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr21_+_29130961 | 1.47 |
ENST00000399925.5
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_+_154405326 | 1.43 |
ENST00000368485.8
|
IL6R
|
interleukin 6 receptor |
| chr1_-_11847772 | 1.43 |
ENST00000376480.7
ENST00000610706.1 |
NPPA
|
natriuretic peptide A |
| chr12_-_18738006 | 1.42 |
ENST00000266505.12
ENST00000543242.5 ENST00000539072.5 ENST00000541966.5 ENST00000648272.1 |
PLCZ1
|
phospholipase C zeta 1 |
| chr7_+_143316105 | 1.37 |
ENST00000343257.7
ENST00000650516.1 |
CLCN1
|
chloride voltage-gated channel 1 |
| chr1_+_154405193 | 1.33 |
ENST00000622330.4
ENST00000344086.8 |
IL6R
|
interleukin 6 receptor |
| chr3_+_167735238 | 1.31 |
ENST00000472941.5
|
SERPINI1
|
serpin family I member 1 |
| chr5_+_139795795 | 1.30 |
ENST00000274710.4
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr1_+_26410809 | 1.30 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr21_+_29130630 | 1.29 |
ENST00000399926.5
ENST00000399928.6 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr1_-_24930263 | 1.26 |
ENST00000308873.11
|
RUNX3
|
RUNX family transcription factor 3 |
| chr7_+_142666272 | 1.21 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2 like 6 |
| chr1_+_154405573 | 1.17 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr3_+_167735704 | 1.16 |
ENST00000446050.7
ENST00000295777.9 ENST00000472747.2 |
SERPINI1
|
serpin family I member 1 |
| chr9_-_97938157 | 1.07 |
ENST00000616898.2
|
HEMGN
|
hemogen |
| chr11_-_128867268 | 1.07 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr11_-_128867364 | 1.07 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr12_-_90955172 | 1.07 |
ENST00000358859.3
|
CCER1
|
coiled-coil glutamate rich protein 1 |
| chr1_+_15736251 | 1.04 |
ENST00000294454.6
|
SLC25A34
|
solute carrier family 25 member 34 |
| chr15_-_38560050 | 1.03 |
ENST00000558432.5
|
RASGRP1
|
RAS guanyl releasing protein 1 |
| chr18_-_58629084 | 0.99 |
ENST00000361673.4
|
ALPK2
|
alpha kinase 2 |
| chrX_+_70423031 | 0.98 |
ENST00000453994.6
ENST00000538649.5 ENST00000536730.5 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr11_+_122838492 | 0.98 |
ENST00000227348.9
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr8_+_55102012 | 0.94 |
ENST00000327381.7
|
XKR4
|
XK related 4 |
| chr5_+_55024250 | 0.91 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chrX_+_71223216 | 0.89 |
ENST00000361726.7
|
GJB1
|
gap junction protein beta 1 |
| chr11_-_16356538 | 0.88 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr7_-_28180735 | 0.88 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chrX_+_124346544 | 0.84 |
ENST00000371139.9
|
SH2D1A
|
SH2 domain containing 1A |
| chr5_-_75511213 | 0.83 |
ENST00000644445.1
ENST00000646302.1 ENST00000644912.1 ENST00000642809.1 ENST00000644377.1 |
CERT1
|
ceramide transporter 1 |
| chr1_-_11848345 | 0.83 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr11_-_85719313 | 0.82 |
ENST00000526999.5
|
SYTL2
|
synaptotagmin like 2 |
| chr22_+_40045451 | 0.82 |
ENST00000402203.5
|
TNRC6B
|
trinucleotide repeat containing adaptor 6B |
| chr7_+_69967464 | 0.81 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr15_+_92393841 | 0.80 |
ENST00000268164.8
ENST00000539113.5 ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr2_+_218607914 | 0.80 |
ENST00000417849.5
|
PLCD4
|
phospholipase C delta 4 |
| chr3_+_134795248 | 0.78 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chrX_+_70423301 | 0.77 |
ENST00000374382.4
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr10_-_16521871 | 0.74 |
ENST00000298943.4
|
C1QL3
|
complement C1q like 3 |
| chrX_+_124346525 | 0.73 |
ENST00000360027.4
|
SH2D1A
|
SH2 domain containing 1A |
| chr10_-_75109085 | 0.71 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr2_+_102473219 | 0.70 |
ENST00000295269.5
|
SLC9A4
|
solute carrier family 9 member A4 |
| chr3_+_134795277 | 0.69 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1 |
| chr1_+_172659095 | 0.69 |
ENST00000367721.3
ENST00000340030.4 |
FASLG
|
Fas ligand |
| chr2_+_209771972 | 0.69 |
ENST00000439458.5
ENST00000272845.10 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr2_+_209771814 | 0.66 |
ENST00000673951.1
ENST00000673920.1 |
UNC80
|
unc-80 homolog, NALCN channel complex subunit |
| chr2_+_218607861 | 0.66 |
ENST00000450993.7
|
PLCD4
|
phospholipase C delta 4 |
| chr10_-_101229449 | 0.66 |
ENST00000370193.4
|
LBX1
|
ladybird homeobox 1 |
| chr8_-_94895195 | 0.66 |
ENST00000308108.9
ENST00000396133.7 |
CCNE2
|
cyclin E2 |
| chr20_+_46029165 | 0.66 |
ENST00000616201.4
ENST00000616202.4 ENST00000616933.4 ENST00000626937.2 |
SLC12A5
|
solute carrier family 12 member 5 |
| chr8_+_737595 | 0.65 |
ENST00000637795.2
|
DLGAP2
|
DLG associated protein 2 |
| chr8_+_55102448 | 0.65 |
ENST00000622811.1
|
XKR4
|
XK related 4 |
| chr5_-_75511596 | 0.64 |
ENST00000643158.1
ENST00000261415.12 ENST00000646713.1 ENST00000643773.1 ENST00000645866.1 ENST00000644072.2 ENST00000643780.2 ENST00000645483.1 ENST00000642556.1 ENST00000646511.1 |
CERT1
|
ceramide transporter 1 |
| chr6_+_104940866 | 0.63 |
ENST00000637759.1
|
LIN28B
|
lin-28 homolog B |
| chr1_-_225889143 | 0.62 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr1_-_37034492 | 0.62 |
ENST00000373091.8
|
GRIK3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr15_+_36702009 | 0.60 |
ENST00000562489.1
|
CDIN1
|
CDAN1 interacting nuclease 1 |
| chr12_-_89630552 | 0.60 |
ENST00000393164.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr1_-_110606009 | 0.60 |
ENST00000640774.2
ENST00000638616.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr12_+_57460127 | 0.59 |
ENST00000532291.5
ENST00000543426.5 ENST00000546141.5 |
GLI1
|
GLI family zinc finger 1 |
| chr10_-_97185507 | 0.59 |
ENST00000314867.9
|
SLIT1
|
slit guidance ligand 1 |
| chrX_+_124346571 | 0.58 |
ENST00000477673.2
|
SH2D1A
|
SH2 domain containing 1A |
| chr2_-_27264083 | 0.58 |
ENST00000432351.5
|
SLC30A3
|
solute carrier family 30 member 3 |
| chr14_-_20590823 | 0.57 |
ENST00000556526.1
|
RNASE12
|
ribonuclease A family member 12 (inactive) |
| chr2_-_2326378 | 0.57 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr17_-_63873676 | 0.56 |
ENST00000613718.3
ENST00000392886.7 ENST00000345366.8 ENST00000336844.9 ENST00000560142.5 |
CSH2
|
chorionic somatomammotropin hormone 2 |
| chr11_-_62921807 | 0.55 |
ENST00000543973.1
|
CHRM1
|
cholinergic receptor muscarinic 1 |
| chr1_+_206507520 | 0.54 |
ENST00000579436.7
|
RASSF5
|
Ras association domain family member 5 |
| chr4_+_87650277 | 0.54 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr8_+_94895763 | 0.53 |
ENST00000523378.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr2_+_176107272 | 0.53 |
ENST00000249504.7
|
HOXD11
|
homeobox D11 |
| chr5_+_150660841 | 0.52 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3 |
| chr15_+_75724034 | 0.51 |
ENST00000332145.3
|
ODF3L1
|
outer dense fiber of sperm tails 3 like 1 |
| chr5_+_83471668 | 0.51 |
ENST00000342785.8
ENST00000343200.9 |
VCAN
|
versican |
| chr2_-_199457931 | 0.50 |
ENST00000417098.6
|
SATB2
|
SATB homeobox 2 |
| chr5_-_79514127 | 0.49 |
ENST00000334082.11
|
HOMER1
|
homer scaffold protein 1 |
| chr12_-_110742929 | 0.49 |
ENST00000340766.9
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr5_+_150661243 | 0.49 |
ENST00000517768.6
|
MYOZ3
|
myozenin 3 |
| chr16_+_2537997 | 0.48 |
ENST00000441549.7
ENST00000268673.11 ENST00000342085.9 ENST00000389224.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase 1 |
| chr1_+_69567906 | 0.48 |
ENST00000651989.2
|
LRRC7
|
leucine rich repeat containing 7 |
| chr11_+_64306227 | 0.47 |
ENST00000405666.5
ENST00000468670.2 |
ESRRA
|
estrogen related receptor alpha |
| chr3_-_187745460 | 0.47 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor |
| chr8_+_22578249 | 0.47 |
ENST00000456545.5
|
PDLIM2
|
PDZ and LIM domain 2 |
| chr1_-_37034129 | 0.46 |
ENST00000373093.4
|
GRIK3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr8_+_32647080 | 0.46 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr2_-_40453438 | 0.46 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr16_-_18926408 | 0.45 |
ENST00000446231.7
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chrX_-_138711663 | 0.45 |
ENST00000315930.11
|
FGF13
|
fibroblast growth factor 13 |
| chr17_-_39401593 | 0.45 |
ENST00000394294.7
ENST00000264658.11 ENST00000583610.5 ENST00000647139.1 |
FBXL20
|
F-box and leucine rich repeat protein 20 |
| chr11_-_27722021 | 0.45 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor |
| chr19_+_35031263 | 0.45 |
ENST00000640135.1
ENST00000596348.2 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr20_+_46029206 | 0.43 |
ENST00000243964.7
|
SLC12A5
|
solute carrier family 12 member 5 |
| chr1_+_206507546 | 0.43 |
ENST00000580449.5
ENST00000581503.6 |
RASSF5
|
Ras association domain family member 5 |
| chr3_-_66500973 | 0.43 |
ENST00000383703.3
ENST00000273261.8 |
LRIG1
|
leucine rich repeats and immunoglobulin like domains 1 |
| chr14_+_52730154 | 0.43 |
ENST00000354586.5
ENST00000442123.6 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr1_+_151612001 | 0.42 |
ENST00000642376.1
ENST00000368843.8 ENST00000458013.7 |
SNX27
|
sorting nexin 27 |
| chr15_+_60004305 | 0.42 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr14_+_93430927 | 0.42 |
ENST00000393151.6
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr6_-_31652115 | 0.42 |
ENST00000454165.1
ENST00000428326.5 ENST00000452994.5 |
BAG6
|
BAG cochaperone 6 |
| chr12_-_116276759 | 0.42 |
ENST00000548743.2
|
MED13L
|
mediator complex subunit 13L |
| chr3_+_63819293 | 0.41 |
ENST00000295896.13
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr4_+_139301478 | 0.41 |
ENST00000296543.10
ENST00000398947.1 |
NAA15
|
N-alpha-acetyltransferase 15, NatA auxiliary subunit |
| chr17_-_63896568 | 0.40 |
ENST00000610991.1
ENST00000316193.13 ENST00000329882.8 |
CSH1
|
chorionic somatomammotropin hormone 1 |
| chr14_+_93430853 | 0.40 |
ENST00000553484.5
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr8_+_32548210 | 0.40 |
ENST00000523079.5
ENST00000650919.1 |
NRG1
|
neuregulin 1 |
| chr6_-_31651920 | 0.39 |
ENST00000434444.5
|
BAG6
|
BAG cochaperone 6 |
| chr8_+_32548303 | 0.39 |
ENST00000650967.1
|
NRG1
|
neuregulin 1 |
| chr2_-_110534010 | 0.39 |
ENST00000437167.1
|
RGPD6
|
RANBP2 like and GRIP domain containing 6 |
| chr15_-_82647123 | 0.39 |
ENST00000569257.5
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_+_111878926 | 0.39 |
ENST00000528125.5
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr11_-_85719111 | 0.39 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr14_+_90398159 | 0.39 |
ENST00000544280.6
|
CALM1
|
calmodulin 1 |
| chr11_-_85719045 | 0.38 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr14_+_78170336 | 0.38 |
ENST00000634499.2
ENST00000335750.7 |
NRXN3
|
neurexin 3 |
| chrX_-_19670983 | 0.38 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr15_+_90906741 | 0.38 |
ENST00000557865.5
|
MAN2A2
|
mannosidase alpha class 2A member 2 |
| chr2_+_37231761 | 0.37 |
ENST00000416653.5
|
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr2_+_37231798 | 0.37 |
ENST00000439218.5
ENST00000432075.1 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr3_-_52409783 | 0.37 |
ENST00000470173.1
ENST00000296288.9 ENST00000460680.6 |
BAP1
|
BRCA1 associated protein 1 |
| chr12_+_21372899 | 0.37 |
ENST00000240652.8
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr16_+_2537979 | 0.36 |
ENST00000566659.1
|
PDPK1
|
3-phosphoinositide dependent protein kinase 1 |
| chr12_-_120368069 | 0.36 |
ENST00000546985.1
|
MSI1
|
musashi RNA binding protein 1 |
| chr14_+_75279961 | 0.36 |
ENST00000557139.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chr5_-_34043205 | 0.36 |
ENST00000382065.8
ENST00000231338.7 |
C1QTNF3
|
C1q and TNF related 3 |
| chr4_-_139301204 | 0.36 |
ENST00000505036.5
ENST00000539002.5 ENST00000544855.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr2_+_60881515 | 0.35 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit |
| chrX_-_139205006 | 0.35 |
ENST00000436198.5
ENST00000626909.2 |
FGF13
|
fibroblast growth factor 13 |
| chr1_+_158355894 | 0.35 |
ENST00000368162.2
|
CD1E
|
CD1e molecule |
| chr14_+_78403686 | 0.35 |
ENST00000553631.1
ENST00000554719.5 |
NRXN3
|
neurexin 3 |
| chr5_+_75512058 | 0.34 |
ENST00000514296.5
|
POLK
|
DNA polymerase kappa |
| chrM_+_10464 | 0.34 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4L |
| chr7_-_150978284 | 0.34 |
ENST00000262186.10
|
KCNH2
|
potassium voltage-gated channel subfamily H member 2 |
| chr2_+_17816458 | 0.33 |
ENST00000281047.4
|
MSGN1
|
mesogenin 1 |
| chr12_-_110742839 | 0.33 |
ENST00000551676.5
ENST00000550991.5 ENST00000335007.10 |
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr22_-_41946729 | 0.32 |
ENST00000402420.1
|
CENPM
|
centromere protein M |
| chr17_-_7329266 | 0.32 |
ENST00000571887.5
ENST00000315614.11 ENST00000399464.7 ENST00000570460.5 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr7_-_72336995 | 0.32 |
ENST00000329008.9
|
CALN1
|
calneuron 1 |
| chr9_-_72364504 | 0.32 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr6_+_49499916 | 0.31 |
ENST00000371197.9
|
GLYATL3
|
glycine-N-acyltransferase like 3 |
| chr2_-_61471062 | 0.31 |
ENST00000398571.7
|
USP34
|
ubiquitin specific peptidase 34 |
| chr1_+_145927105 | 0.31 |
ENST00000437797.5
ENST00000601726.3 ENST00000599626.5 ENST00000599147.5 ENST00000595494.5 ENST00000595518.5 ENST00000597144.5 ENST00000599469.5 ENST00000598354.5 ENST00000598103.5 ENST00000600340.5 ENST00000630257.2 ENST00000625258.1 |
LIX1L-AS1
ENSG00000280778.1
|
LIX1L antisense RNA 1 novel protein, lncRNA-POLR3GL readthrough |
| chr11_-_33869816 | 0.30 |
ENST00000395833.7
|
LMO2
|
LIM domain only 2 |
| chrX_+_110002635 | 0.30 |
ENST00000372072.7
|
TMEM164
|
transmembrane protein 164 |
| chr11_+_73647645 | 0.30 |
ENST00000545798.5
ENST00000539157.5 ENST00000546251.5 ENST00000535582.5 ENST00000538227.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr11_-_33892010 | 0.30 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chr11_+_61680373 | 0.30 |
ENST00000257215.10
|
DAGLA
|
diacylglycerol lipase alpha |
| chr17_-_63918817 | 0.29 |
ENST00000458650.6
ENST00000351388.8 ENST00000342364.8 ENST00000617086.1 ENST00000323322.10 ENST00000392824.8 |
GH1
CSHL1
|
growth hormone 1 chorionic somatomammotropin hormone like 1 |
| chr11_-_132943732 | 0.29 |
ENST00000374778.4
|
OPCML
|
opioid binding protein/cell adhesion molecule like |
| chr11_+_17547259 | 0.29 |
ENST00000399397.6
ENST00000399391.7 |
OTOG
|
otogelin |
| chr6_-_31651964 | 0.29 |
ENST00000433828.5
ENST00000456286.5 |
BAG6
|
BAG cochaperone 6 |
| chr12_+_113185702 | 0.29 |
ENST00000548278.2
|
RITA1
|
RBPJ interacting and tubulin associated 1 |
| chr2_-_25252251 | 0.29 |
ENST00000380746.8
ENST00000402667.1 |
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr8_+_122781621 | 0.29 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr11_-_85719160 | 0.29 |
ENST00000389958.7
ENST00000527794.5 |
SYTL2
|
synaptotagmin like 2 |
| chr4_-_113517607 | 0.29 |
ENST00000509594.1
|
CAMK2D
|
calcium/calmodulin dependent protein kinase II delta |
| chrM_+_10759 | 0.29 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 |
| chr7_+_27242796 | 0.28 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr3_+_107645836 | 0.28 |
ENST00000413213.5
|
BBX
|
BBX high mobility group box domain containing |
| chr2_+_60881553 | 0.28 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit |
| chr3_+_107645922 | 0.28 |
ENST00000449271.5
ENST00000425868.5 ENST00000449213.5 |
BBX
|
BBX high mobility group box domain containing |
| chr11_-_114400417 | 0.27 |
ENST00000325636.8
ENST00000623205.2 |
C11orf71
|
chromosome 11 open reading frame 71 |
| chr15_-_82647503 | 0.27 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_+_94021346 | 0.27 |
ENST00000315765.10
|
HEPHL1
|
hephaestin like 1 |
| chrM_+_10055 | 0.27 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chr4_-_101347471 | 0.27 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chrX_+_47191399 | 0.27 |
ENST00000412206.5
ENST00000427561.5 |
UBA1
|
ubiquitin like modifier activating enzyme 1 |
| chr4_-_44448796 | 0.27 |
ENST00000360029.4
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr10_+_7818497 | 0.26 |
ENST00000344293.6
|
TAF3
|
TATA-box binding protein associated factor 3 |
| chr1_-_182672232 | 0.26 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr15_-_82806054 | 0.26 |
ENST00000541889.1
ENST00000334574.12 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr5_+_140107777 | 0.25 |
ENST00000505703.2
ENST00000651386.1 |
PURA
|
purine rich element binding protein A |
| chr10_+_93073873 | 0.25 |
ENST00000224356.5
|
CYP26A1
|
cytochrome P450 family 26 subfamily A member 1 |
| chr3_+_63819368 | 0.25 |
ENST00000616659.1
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chr4_-_23881282 | 0.25 |
ENST00000613098.4
|
PPARGC1A
|
PPARG coactivator 1 alpha |
| chr11_-_74949079 | 0.24 |
ENST00000528219.5
ENST00000684022.1 ENST00000531852.5 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr11_-_18322178 | 0.24 |
ENST00000531848.1
|
HPS5
|
HPS5 biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr11_-_74949114 | 0.23 |
ENST00000527087.5
|
XRRA1
|
X-ray radiation resistance associated 1 |
| chr10_+_113125536 | 0.23 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr15_-_89751292 | 0.22 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1 |
| chr11_-_74949020 | 0.22 |
ENST00000525407.5
|
XRRA1
|
X-ray radiation resistance associated 1 |
| chr11_+_73647701 | 0.21 |
ENST00000543524.5
|
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr7_-_72337601 | 0.21 |
ENST00000446128.1
|
CALN1
|
calneuron 1 |
| chr9_-_114930508 | 0.21 |
ENST00000223795.3
ENST00000618336.4 |
TNFSF8
|
TNF superfamily member 8 |
| chr7_+_27242700 | 0.20 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1 |
| chr12_-_119803383 | 0.20 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr2_-_25252072 | 0.19 |
ENST00000683760.1
|
DNMT3A
|
DNA methyltransferase 3 alpha |
| chr2_+_37231633 | 0.19 |
ENST00000002125.9
ENST00000336237.10 ENST00000431821.5 |
NDUFAF7
|
NADH:ubiquinone oxidoreductase complex assembly factor 7 |
| chr11_-_18322122 | 0.19 |
ENST00000349215.8
ENST00000396253.7 ENST00000438420.6 |
HPS5
|
HPS5 biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr20_+_46029009 | 0.19 |
ENST00000608944.5
|
SLC12A5
|
solute carrier family 12 member 5 |
| chr4_-_101347492 | 0.19 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chrX_+_123962040 | 0.18 |
ENST00000435215.5
|
STAG2
|
stromal antigen 2 |
| chr4_-_185811088 | 0.18 |
ENST00000421639.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_111879154 | 0.17 |
ENST00000260257.9
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr16_+_57106021 | 0.16 |
ENST00000566259.1
|
CPNE2
|
copine 2 |
| chr17_-_45425620 | 0.16 |
ENST00000376922.6
|
ARHGAP27
|
Rho GTPase activating protein 27 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.5 | 1.5 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.4 | 2.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.3 | 1.3 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.3 | 1.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.2 | 0.6 | GO:0060032 | notochord regression(GO:0060032) |
| 0.2 | 0.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 1.0 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.9 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.5 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 1.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.9 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.8 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.3 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.5 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.1 | GO:1904327 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.5 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.6 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 1.4 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.2 | GO:2000184 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) response to resveratrol(GO:1904638) cellular response to resveratrol(GO:1904639) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.1 | 2.6 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 | 0.3 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.2 | GO:0008078 | mesodermal cell migration(GO:0008078) cardiac cell fate determination(GO:0060913) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.1 | 0.4 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 1.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.8 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 3.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.5 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 1.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 1.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.2 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.0 | 0.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.8 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.6 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.9 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.4 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 2.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.4 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 2.5 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.7 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.4 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.4 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 0.7 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 1.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.2 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.1 | 3.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 4.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.2 | 1.8 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 0.6 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.2 | 2.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 0.9 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 1.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.4 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 1.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 3.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.1 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.8 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |