Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for TEAD3_TEAD1
Z-value: 7.56
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
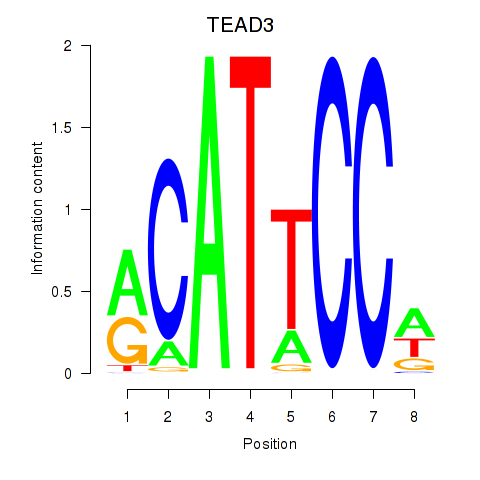
TEAD3
|
ENSG00000007866.22 | TEAD3 |
|
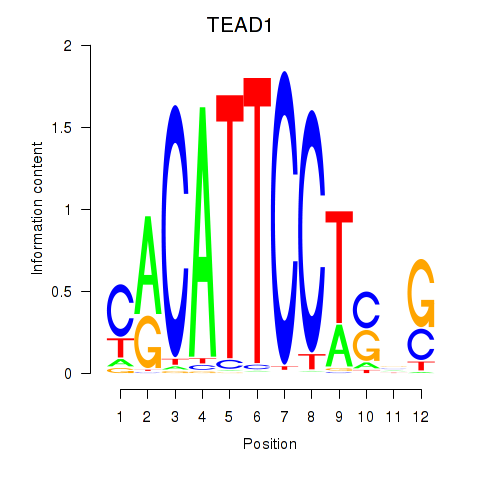
TEAD1
|
ENSG00000187079.20 | TEAD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD1 | hg38_v1_chr11_+_12674397_12674442 | 0.91 | 4.2e-13 | Click! |
| TEAD3 | hg38_v1_chr6_-_35497042_35497117 | 0.80 | 3.7e-08 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.2 | 68.6 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 12.0 | 47.8 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 7.6 | 45.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 5.8 | 17.3 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 4.4 | 17.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 4.4 | 8.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 4.1 | 20.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 4.0 | 15.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 3.9 | 11.7 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 3.6 | 25.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 3.4 | 20.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 3.1 | 18.7 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 3.0 | 73.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.9 | 8.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 2.9 | 20.2 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 2.9 | 5.8 | GO:0072190 | ureter urothelium development(GO:0072190) ureter morphogenesis(GO:0072197) |
| 2.6 | 7.9 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 2.5 | 14.9 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.4 | 46.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 2.4 | 7.1 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 2.3 | 9.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 2.3 | 6.8 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 2.2 | 8.9 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 2.2 | 10.8 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 2.1 | 12.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 2.1 | 6.4 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 2.1 | 6.2 | GO:1901491 | axial mesoderm formation(GO:0048320) negative regulation of lymphangiogenesis(GO:1901491) |
| 2.1 | 14.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.9 | 18.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.9 | 5.6 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 1.8 | 27.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.8 | 8.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.8 | 40.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.7 | 19.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.7 | 10.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.7 | 6.7 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 1.7 | 36.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 1.6 | 8.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.6 | 17.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 1.6 | 23.7 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.6 | 6.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.6 | 6.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 1.5 | 5.9 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 1.4 | 8.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.4 | 5.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.4 | 62.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 1.3 | 6.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 1.3 | 3.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 1.2 | 3.7 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 1.2 | 13.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 1.2 | 1.2 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 1.2 | 3.5 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.1 | 17.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 1.1 | 1.1 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 1.0 | 7.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.0 | 5.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.0 | 11.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.0 | 56.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 1.0 | 10.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.9 | 5.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.9 | 6.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 18.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.9 | 6.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.9 | 2.7 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.9 | 5.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.9 | 3.4 | GO:0022012 | subpallium cell proliferation in forebrain(GO:0022012) lateral ganglionic eminence cell proliferation(GO:0022018) lambdoid suture morphogenesis(GO:0060366) sagittal suture morphogenesis(GO:0060367) anterior semicircular canal development(GO:0060873) lateral semicircular canal development(GO:0060875) |
| 0.9 | 5.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.8 | 10.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.8 | 19.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.8 | 3.9 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.8 | 5.4 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.7 | 27.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.7 | 11.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.7 | 4.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.7 | 4.9 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.7 | 4.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.7 | 4.8 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.6 | 5.7 | GO:0072093 | metanephric renal vesicle formation(GO:0072093) |
| 0.6 | 7.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.6 | 4.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.6 | 7.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.6 | 12.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.6 | 30.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.6 | 10.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.6 | 5.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.6 | 4.1 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.6 | 28.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.6 | 3.4 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 5.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.6 | 11.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.5 | 9.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.5 | 1.6 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.5 | 2.2 | GO:0035853 | chromosome passenger complex localization to spindle midzone(GO:0035853) |
| 0.5 | 3.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.5 | 18.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.5 | 3.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 1.6 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.5 | 3.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.5 | 10.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.5 | 4.0 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.5 | 14.0 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.5 | 11.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.5 | 11.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.5 | 5.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.5 | 7.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.5 | 13.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.5 | 7.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.5 | 14.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.5 | 3.3 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.5 | 0.5 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) positive regulation of miRNA metabolic process(GO:2000630) |
| 0.5 | 5.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 | 5.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.3 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.4 | 6.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.4 | 2.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.4 | 3.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.4 | 1.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 7.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.4 | 1.7 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.4 | 3.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 2.9 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.4 | 3.7 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.4 | 3.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 1.2 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.4 | 4.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.4 | 1.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.4 | 1.6 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.4 | 7.0 | GO:1901626 | regulation of postsynaptic membrane organization(GO:1901626) |
| 0.4 | 6.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 1.5 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.4 | 3.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.4 | 55.7 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.4 | 2.9 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.4 | 1.4 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.4 | 9.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 0.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.3 | 3.7 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.3 | 1.3 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 2.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 1.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.3 | 0.6 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.3 | 8.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.3 | 6.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.3 | 17.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 20.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.3 | 16.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.3 | 1.8 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.3 | 2.4 | GO:0008354 | germ cell migration(GO:0008354) regulation of collagen catabolic process(GO:0010710) |
| 0.3 | 2.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 44.4 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.3 | 4.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 2.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.3 | 3.7 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.3 | 13.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.3 | 1.1 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 6.6 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.3 | 2.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 5.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 8.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 5.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 2.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 2.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.2 | 1.2 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.2 | 3.8 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 4.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 2.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 32.5 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.2 | 5.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 96.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.2 | 1.8 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.2 | 6.1 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 11.0 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.2 | 3.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 16.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 1.5 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 5.6 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.2 | 2.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.2 | 2.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.8 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 2.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 12.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.2 | 4.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 0.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 2.7 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 28.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.2 | 6.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 0.5 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.5 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.2 | 3.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 3.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 2.0 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 2.8 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.2 | 1.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.2 | 1.6 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 0.6 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.2 | 3.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.2 | 1.2 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 7.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 13.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 3.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 3.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.7 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 1.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 3.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 4.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 5.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 7.0 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 3.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 6.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 4.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 16.6 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 1.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 10.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 4.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 12.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.5 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 2.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 4.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.3 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.9 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 2.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 1.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 1.0 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.4 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 1.0 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 3.9 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 9.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.8 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 2.9 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 2.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 2.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 5.7 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0071316 | olfactory nerve development(GO:0021553) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) cellular response to nicotine(GO:0071316) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 2.3 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 1.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.4 | GO:1903363 | negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 4.4 | 30.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 4.2 | 72.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.8 | 18.8 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 3.4 | 67.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 3.3 | 9.8 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 3.1 | 9.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 2.7 | 5.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 2.6 | 7.8 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 2.6 | 119.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 2.3 | 35.1 | GO:0030478 | actin cap(GO:0030478) |
| 2.1 | 6.4 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.0 | 7.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.7 | 8.5 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.7 | 5.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.5 | 5.8 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.4 | 5.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 54.6 | GO:0036379 | myofilament(GO:0036379) |
| 1.2 | 6.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 1.2 | 17.9 | GO:0043256 | laminin complex(GO:0043256) |
| 1.1 | 42.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 8.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 28.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.8 | 5.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 64.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.7 | 7.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.7 | 11.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.6 | 19.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.6 | 5.7 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.6 | 33.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.6 | 161.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.5 | 9.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.5 | 14.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 3.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 5.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 24.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 8.9 | GO:0031672 | A band(GO:0031672) |
| 0.4 | 7.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 3.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 5.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.4 | 1.1 | GO:0097135 | X chromosome(GO:0000805) cyclin E2-CDK2 complex(GO:0097135) |
| 0.4 | 4.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 3.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.3 | 1.0 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 4.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 6.4 | GO:0031674 | I band(GO:0031674) |
| 0.3 | 3.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 6.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 2.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.3 | 5.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 5.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.3 | 6.0 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 3.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 10.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 1.0 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 1.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 22.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 26.4 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 4.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 44.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 2.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 45.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 3.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 105.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 0.6 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 16.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 3.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 4.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 3.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 16.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 5.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 18.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 84.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 14.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.5 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 17.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 8.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 3.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 26.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 7.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 5.6 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 6.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 6.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 10.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.3 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 6.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 24.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 10.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 8.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0071004 | U1 snRNP(GO:0005685) U2-type prespliceosome(GO:0071004) |
| 0.0 | 2.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 13.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 14.8 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 26.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 6.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.2 | 68.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 6.6 | 26.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.8 | 11.4 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 3.1 | 18.9 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 2.9 | 17.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 2.8 | 8.5 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 2.8 | 25.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 2.5 | 76.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 2.2 | 67.8 | GO:0031005 | filamin binding(GO:0031005) |
| 2.1 | 6.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 2.1 | 6.4 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 2.0 | 16.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.9 | 18.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.8 | 43.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.6 | 7.9 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 1.5 | 196.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.5 | 13.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.5 | 5.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.4 | 36.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 1.3 | 7.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.3 | 12.8 | GO:0031432 | titin binding(GO:0031432) |
| 1.2 | 7.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 1.2 | 4.9 | GO:0030379 | neurotensin receptor activity, non-G-protein coupled(GO:0030379) |
| 1.2 | 8.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.2 | 38.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.2 | 17.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.2 | 18.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 1.1 | 10.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.0 | 18.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.0 | 4.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.0 | 4.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.9 | 8.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.9 | 5.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.9 | 38.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.8 | 10.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.8 | 37.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.8 | 8.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.7 | 2.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.7 | 3.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 2.1 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.7 | 13.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.6 | 9.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.6 | 7.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 13.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.6 | 18.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.6 | 8.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 3.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 1.6 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.5 | 2.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 1.6 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.5 | 6.7 | GO:0036122 | BMP binding(GO:0036122) |
| 0.5 | 7.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 7.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 13.8 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.5 | 16.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 4.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 15.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 6.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 9.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 1.5 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.4 | 3.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 3.0 | GO:0044020 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.4 | 10.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 16.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 4.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 9.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.3 | 11.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 12.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 6.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.3 | 9.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 9.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 1.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.3 | 1.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 38.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 34.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 4.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 3.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 7.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 4.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 3.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 8.7 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 7.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 20.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 7.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 4.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 2.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 5.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 1.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 4.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 31.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.2 | 14.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 9.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 8.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 3.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 5.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 1.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 24.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 14.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 3.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 2.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 2.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 10.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 1.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 3.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 4.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 4.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 31.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 3.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 58.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 3.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 2.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.7 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 28.8 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 15.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 11.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 44.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 2.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 4.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 3.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 17.1 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 10.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 3.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 6.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 22.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 22.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 6.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 1.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 4.9 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 2.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 9.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 4.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 4.4 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 2.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 1.2 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 10.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 3.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 6.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 74.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 1.2 | 27.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 17.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 15.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 23.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 38.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 31.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 27.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 39.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 31.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.4 | 6.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 2.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 4.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 3.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 17.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 37.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 40.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 5.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 33.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.3 | 4.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 11.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 8.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 22.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 17.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 31.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 11.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 5.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 12.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 13.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 5.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 8.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 4.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 3.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 8.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 8.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 19.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 3.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 3.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 11.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 9.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 89.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 2.1 | 84.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 2.0 | 127.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.5 | 86.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.8 | 33.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.7 | 28.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.6 | 26.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 18.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.5 | 21.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 13.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.5 | 10.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.5 | 6.9 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.4 | 37.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.4 | 1.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 6.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 2.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 9.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 29.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 11.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 5.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 32.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 8.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 8.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 13.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 6.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 6.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 16.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 25.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 9.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 9.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 2.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 7.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 6.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 5.5 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 3.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.9 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |