Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for VENTX
Z-value: 1.64
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.8 | VENTX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VENTX | hg38_v1_chr10_+_133237849_133237863 | 0.19 | 2.9e-01 | Click! |
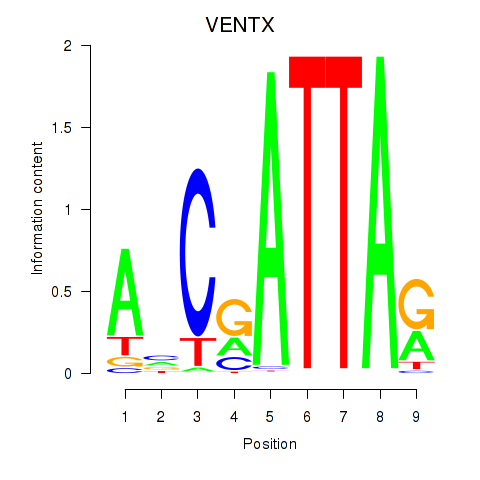
Activity profile of VENTX motif
Sorted Z-values of VENTX motif
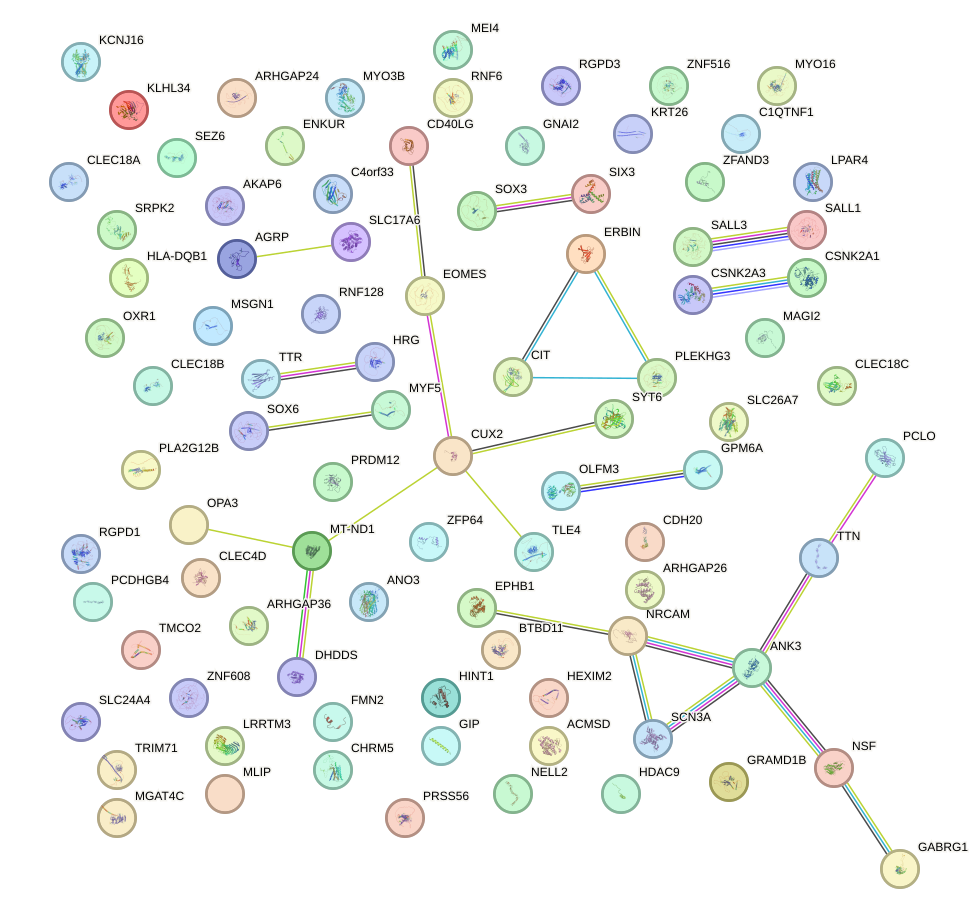
Network of associatons between targets according to the STRING database.
First level regulatory network of VENTX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_154590735 | 4.71 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr12_+_107318395 | 3.32 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr18_+_31591869 | 3.25 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr3_+_186666003 | 2.36 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein |
| chr16_-_51151259 | 2.28 |
ENST00000251020.9
|
SALL1
|
spalt like transcription factor 1 |
| chr2_+_134838610 | 2.23 |
ENST00000356140.10
ENST00000392928.5 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr17_+_46624036 | 1.99 |
ENST00000575068.5
|
NSF
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr14_+_21965451 | 1.96 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chr14_+_92323154 | 1.95 |
ENST00000532405.6
ENST00000676001.1 ENST00000531433.5 |
SLC24A4
|
solute carrier family 24 member 4 |
| chr8_+_49911604 | 1.81 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr5_+_142874881 | 1.78 |
ENST00000475287.2
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr11_+_123560240 | 1.76 |
ENST00000529432.5
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr16_-_51151238 | 1.65 |
ENST00000566102.1
|
SALL1
|
spalt like transcription factor 1 |
| chr18_+_61333424 | 1.59 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chrX_+_78747705 | 1.51 |
ENST00000614823.5
ENST00000435339.3 ENST00000514744.5 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr3_+_32817990 | 1.51 |
ENST00000383763.6
|
TRIM71
|
tripartite motif containing 71 |
| chr12_+_111034136 | 1.50 |
ENST00000261726.11
|
CUX2
|
cut like homeobox 2 |
| chr7_+_54542300 | 1.45 |
ENST00000302287.7
ENST00000407838.7 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr8_+_91249307 | 1.40 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr17_-_31318818 | 1.35 |
ENST00000578584.5
|
ENSG00000265118.5
|
novel protein |
| chrX_+_136648138 | 1.34 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr11_+_22338333 | 1.34 |
ENST00000263160.4
|
SLC17A6
|
solute carrier family 17 member 6 |
| chr3_+_134795248 | 1.34 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chr7_+_54542362 | 1.32 |
ENST00000402613.4
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr12_-_119804298 | 1.29 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chrX_+_106920393 | 1.28 |
ENST00000336803.2
|
CLDN2
|
claudin 2 |
| chr2_+_86907953 | 1.25 |
ENST00000409776.6
|
RGPD1
|
RANBP2 like and GRIP domain containing 1 |
| chr12_-_119803383 | 1.24 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr16_+_70173783 | 1.21 |
ENST00000541793.7
ENST00000314151.12 ENST00000565806.5 ENST00000569347.6 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18 member C |
| chr7_+_54542393 | 1.18 |
ENST00000404951.5
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr17_+_70104848 | 1.14 |
ENST00000392670.5
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr6_+_77653035 | 1.13 |
ENST00000684080.1
|
MEI4
|
meiotic double-stranded break formation protein 4 |
| chr7_-_83162899 | 1.13 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr8_+_49911396 | 1.08 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr16_-_74421392 | 1.08 |
ENST00000339953.9
ENST00000620745.1 ENST00000682950.1 |
CLEC18B
|
C-type lectin domain family 18 member B |
| chr17_+_70104991 | 1.07 |
ENST00000587698.5
ENST00000587892.1 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chrX_+_131058340 | 1.07 |
ENST00000276211.10
ENST00000370922.5 |
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr2_+_170178136 | 1.05 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr3_+_134795277 | 1.05 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1 |
| chr1_+_40247926 | 1.04 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr14_+_64704380 | 1.03 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr10_-_72954790 | 1.01 |
ENST00000373032.4
|
PLA2G12B
|
phospholipase A2 group XIIB |
| chr4_-_46124046 | 1.01 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr7_-_108240049 | 0.99 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr15_+_33968484 | 0.98 |
ENST00000383263.7
|
CHRM5
|
cholinergic receptor muscarinic 5 |
| chr16_-_67483541 | 0.98 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chrX_+_106693838 | 0.96 |
ENST00000324342.7
|
RNF128
|
ring finger protein 128 |
| chr12_+_69825221 | 0.96 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chrX_+_136648214 | 0.96 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr14_+_32329341 | 0.95 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr9_-_23826231 | 0.94 |
ENST00000397312.7
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr16_+_69951180 | 0.94 |
ENST00000288040.10
ENST00000449317.6 |
CLEC18A
|
C-type lectin domain family 18 member A |
| chr4_-_175812236 | 0.91 |
ENST00000505375.5
|
GPM6A
|
glycoprotein M6A |
| chr10_+_66926028 | 0.90 |
ENST00000361320.5
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3 |
| chr2_-_165203870 | 0.89 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr14_+_22202561 | 0.87 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr3_-_27722699 | 0.87 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr16_+_69950907 | 0.86 |
ENST00000393701.6
ENST00000568461.5 |
CLEC18A
|
C-type lectin domain family 18 member A |
| chr1_-_101846957 | 0.85 |
ENST00000338858.9
|
OLFM3
|
olfactomedin 3 |
| chr19_-_45584810 | 0.85 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr1_-_114153863 | 0.82 |
ENST00000610222.3
ENST00000369547.6 ENST00000641643.2 |
SYT6
|
synaptotagmin 6 |
| chrX_-_21658324 | 0.82 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr10_-_60572599 | 0.82 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr1_+_240123148 | 0.81 |
ENST00000681824.1
|
FMN2
|
formin 2 |
| chr11_-_16356538 | 0.80 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr12_-_44875468 | 0.78 |
ENST00000553120.1
|
NELL2
|
neural EGFL like 2 |
| chr6_+_37929959 | 0.78 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr9_+_79572572 | 0.77 |
ENST00000435650.5
ENST00000414465.5 ENST00000376537.8 |
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr12_+_69825273 | 0.76 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr12_+_48328980 | 0.76 |
ENST00000335017.1
|
H1-7
|
H1.7 linker histone |
| chr1_+_240123121 | 0.75 |
ENST00000681210.1
|
FMN2
|
formin 2 |
| chr10_-_25062279 | 0.75 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr14_+_21841182 | 0.73 |
ENST00000390433.1
|
TRAV12-1
|
T cell receptor alpha variable 12-1 |
| chr11_+_26473900 | 0.72 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr14_+_64715677 | 0.71 |
ENST00000634379.2
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr17_-_40772162 | 0.71 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr14_+_32329256 | 0.71 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr8_+_31639222 | 0.71 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr13_+_108629605 | 0.70 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr13_+_75804270 | 0.70 |
ENST00000447038.5
|
LMO7
|
LIM domain 7 |
| chr8_+_31639291 | 0.69 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chr13_+_75804169 | 0.69 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr12_+_25052220 | 0.67 |
ENST00000550945.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr5_-_124745315 | 0.67 |
ENST00000306315.9
|
ZNF608
|
zinc finger protein 608 |
| chr2_-_178804623 | 0.66 |
ENST00000359218.10
ENST00000342175.11 |
TTN
|
titin |
| chr12_+_8513499 | 0.66 |
ENST00000299665.3
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr6_-_32668368 | 0.65 |
ENST00000399084.5
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr5_-_157059109 | 0.64 |
ENST00000523175.6
ENST00000522693.5 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr12_+_80716906 | 0.64 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr17_+_45161864 | 0.62 |
ENST00000589230.6
ENST00000591576.5 ENST00000591070.6 ENST00000592695.1 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr20_-_52105644 | 0.62 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr4_+_85827745 | 0.61 |
ENST00000509300.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_+_232520384 | 0.61 |
ENST00000617714.2
|
PRSS56
|
serine protease 56 |
| chr12_+_25052512 | 0.60 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr5_+_65926644 | 0.59 |
ENST00000511297.5
ENST00000506030.5 |
ERBIN
|
erbb2 interacting protein |
| chr7_-_105269007 | 0.59 |
ENST00000357311.7
|
SRPK2
|
SRSF protein kinase 2 |
| chr13_-_26221703 | 0.58 |
ENST00000381570.7
ENST00000346166.7 |
RNF6
|
ring finger protein 6 |
| chr10_+_112283399 | 0.57 |
ENST00000643850.1
ENST00000646139.2 ENST00000645243.1 |
TECTB
|
tectorin beta |
| chr19_-_45584769 | 0.57 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chrM_+_3298 | 0.55 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1 |
| chr4_+_129096144 | 0.55 |
ENST00000425929.6
ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr18_+_78979811 | 0.54 |
ENST00000537592.7
|
SALL3
|
spalt like transcription factor 3 |
| chr8_-_42501224 | 0.54 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chrX_-_140505058 | 0.53 |
ENST00000370536.5
|
SOX3
|
SRY-box transcription factor 3 |
| chr22_-_40533808 | 0.53 |
ENST00000422851.1
ENST00000651694.1 ENST00000652095.2 |
MRTFA
|
myocardin related transcription factor A |
| chr12_-_86256299 | 0.53 |
ENST00000552808.6
ENST00000547225.5 |
MGAT4C
|
MGAT4 family member C |
| chr5_-_131165231 | 0.52 |
ENST00000675100.1
ENST00000304043.10 ENST00000513012.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr16_-_74421756 | 0.52 |
ENST00000617101.4
|
CLEC18B
|
C-type lectin domain family 18 member B |
| chr2_+_44941695 | 0.51 |
ENST00000260653.5
|
SIX3
|
SIX homeobox 3 |
| chr6_+_54099565 | 0.50 |
ENST00000511678.5
|
MLIP
|
muscular LMNA interacting protein |
| chr3_-_127288044 | 0.50 |
ENST00000465482.3
|
PRR20G
|
proline rich 20G |
| chr9_+_130664594 | 0.50 |
ENST00000253008.3
ENST00000676323.1 |
PRDM12
|
PR/SET domain 12 |
| chr17_-_29005913 | 0.49 |
ENST00000442608.7
ENST00000317338.17 ENST00000335960.10 |
SEZ6
|
seizure related 6 homolog |
| chr12_+_25052694 | 0.48 |
ENST00000554942.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr14_+_64704102 | 0.48 |
ENST00000555982.5
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr8_+_106726012 | 0.47 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr17_-_48968587 | 0.47 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr1_+_40709475 | 0.47 |
ENST00000372651.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr5_-_131165272 | 0.47 |
ENST00000675491.1
ENST00000506908.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr7_-_78771058 | 0.47 |
ENST00000628781.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr13_+_75804221 | 0.47 |
ENST00000489941.6
ENST00000525373.5 |
LMO7
|
LIM domain 7 |
| chr11_-_11353241 | 0.46 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr18_-_76495191 | 0.46 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr2_+_17816458 | 0.45 |
ENST00000281047.4
|
MSGN1
|
mesogenin 1 |
| chr7_+_18495697 | 0.45 |
ENST00000413380.5
ENST00000430454.5 |
HDAC9
|
histone deacetylase 9 |
| chr12_-_101210232 | 0.44 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8 |
| chr11_-_3837858 | 0.44 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chrX_+_106693751 | 0.43 |
ENST00000418562.5
|
RNF128
|
ring finger protein 128 |
| chr3_+_9792495 | 0.41 |
ENST00000498623.6
|
ARPC4
|
actin related protein 2/3 complex subunit 4 |
| chr7_-_78771108 | 0.41 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_130344837 | 0.41 |
ENST00000485477.5
ENST00000431780.6 |
CPA5
|
carboxypeptidase A5 |
| chr6_+_54099538 | 0.39 |
ENST00000447836.6
|
MLIP
|
muscular LMNA interacting protein |
| chr7_+_130344810 | 0.39 |
ENST00000497503.5
ENST00000463587.5 ENST00000461828.5 ENST00000474905.6 ENST00000494311.1 ENST00000466363.6 |
CPA5
|
carboxypeptidase A5 |
| chr12_+_25052732 | 0.39 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr4_-_121227461 | 0.38 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr12_+_25052634 | 0.37 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr9_-_146140 | 0.37 |
ENST00000475990.5
|
CBWD1
|
COBW domain containing 1 |
| chr4_+_168497113 | 0.37 |
ENST00000511948.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr10_+_84194527 | 0.37 |
ENST00000623527.4
|
CDHR1
|
cadherin related family member 1 |
| chr2_-_202911621 | 0.36 |
ENST00000261015.5
|
WDR12
|
WD repeat domain 12 |
| chr14_+_22482287 | 0.36 |
ENST00000390484.1
|
TRAJ54
|
T cell receptor alpha joining 54 |
| chr9_+_133636355 | 0.36 |
ENST00000393056.8
|
DBH
|
dopamine beta-hydroxylase |
| chr18_+_78980181 | 0.35 |
ENST00000616649.4
ENST00000575389.6 |
SALL3
|
spalt like transcription factor 3 |
| chr11_+_46701010 | 0.34 |
ENST00000311764.3
|
ZNF408
|
zinc finger protein 408 |
| chr10_+_92834594 | 0.34 |
ENST00000371552.8
|
EXOC6
|
exocyst complex component 6 |
| chr7_+_90709231 | 0.34 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr3_-_157503574 | 0.33 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr1_+_244352627 | 0.33 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr4_+_129096113 | 0.32 |
ENST00000508673.5
|
C4orf33
|
chromosome 4 open reading frame 33 |
| chr7_+_151048526 | 0.32 |
ENST00000349064.10
|
ASIC3
|
acid sensing ion channel subunit 3 |
| chr6_+_47781982 | 0.32 |
ENST00000489301.6
ENST00000638973.1 ENST00000371211.6 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr14_+_64986846 | 0.31 |
ENST00000246166.3
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr6_-_138499487 | 0.31 |
ENST00000343505.9
|
NHSL1
|
NHS like 1 |
| chr22_-_30574572 | 0.31 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr8_+_49911801 | 0.30 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr14_+_23240346 | 0.30 |
ENST00000430154.6
|
RNF212B
|
ring finger protein 212B |
| chr4_+_85827891 | 0.30 |
ENST00000514229.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr9_+_133636378 | 0.30 |
ENST00000263611.3
|
DBH
|
dopamine beta-hydroxylase |
| chr5_-_80654956 | 0.30 |
ENST00000439211.7
|
DHFR
|
dihydrofolate reductase |
| chr7_-_151210488 | 0.29 |
ENST00000644661.2
|
H2BE1
|
H2B.E variant histone 1 |
| chr1_+_248508073 | 0.29 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr7_+_90709530 | 0.29 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr7_+_90709816 | 0.29 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14 |
| chr12_-_86256267 | 0.28 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr4_-_83114715 | 0.28 |
ENST00000426923.2
ENST00000311507.9 ENST00000509973.5 |
PLAC8
|
placenta associated 8 |
| chr8_+_106726115 | 0.28 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr2_+_168901290 | 0.28 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr12_-_52434363 | 0.28 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr12_-_52618559 | 0.27 |
ENST00000305748.7
|
KRT73
|
keratin 73 |
| chr3_-_57199938 | 0.26 |
ENST00000473921.2
ENST00000295934.8 |
HESX1
|
HESX homeobox 1 |
| chr15_-_32888239 | 0.26 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr3_-_71360753 | 0.26 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chr18_-_26657401 | 0.25 |
ENST00000580191.5
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr1_+_174447944 | 0.24 |
ENST00000367685.5
|
GPR52
|
G protein-coupled receptor 52 |
| chr8_-_102412686 | 0.24 |
ENST00000220959.8
ENST00000520539.6 |
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr14_+_74239447 | 0.24 |
ENST00000261980.3
|
VSX2
|
visual system homeobox 2 |
| chr11_+_59436469 | 0.23 |
ENST00000641045.1
|
OR5A1
|
olfactory receptor family 5 subfamily A member 1 |
| chr4_+_122239965 | 0.22 |
ENST00000446180.5
|
KIAA1109
|
KIAA1109 |
| chr13_-_103066411 | 0.22 |
ENST00000245312.5
|
SLC10A2
|
solute carrier family 10 member 2 |
| chr12_+_64405046 | 0.22 |
ENST00000540203.5
|
XPOT
|
exportin for tRNA |
| chrX_+_123963164 | 0.21 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr6_+_101398788 | 0.21 |
ENST00000369138.5
ENST00000413795.5 ENST00000358361.7 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr3_+_112041920 | 0.21 |
ENST00000617607.4
|
TMPRSS7
|
transmembrane serine protease 7 |
| chr11_-_27722021 | 0.20 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor |
| chr1_+_42825548 | 0.20 |
ENST00000372514.7
|
ERMAP
|
erythroblast membrane associated protein (Scianna blood group) |
| chr16_-_57946602 | 0.19 |
ENST00000564654.1
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1 |
| chr17_-_50707855 | 0.19 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr11_+_124566660 | 0.19 |
ENST00000641670.1
|
OR8A1
|
olfactory receptor family 8 subfamily A member 1 |
| chr12_-_89352487 | 0.19 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr6_-_49713521 | 0.19 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2 |
| chr7_+_18496269 | 0.19 |
ENST00000432645.6
|
HDAC9
|
histone deacetylase 9 |
| chr12_+_56080126 | 0.18 |
ENST00000411731.6
|
ERBB3
|
erb-b2 receptor tyrosine kinase 3 |
| chr4_+_112860912 | 0.17 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr12_+_52301833 | 0.16 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr4_+_112860981 | 0.16 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr1_+_40709316 | 0.16 |
ENST00000372652.5
|
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr11_-_124320197 | 0.15 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr10_-_54801262 | 0.15 |
ENST00000373965.6
ENST00000616114.4 ENST00000621708.4 ENST00000495484.5 ENST00000395440.5 ENST00000395442.5 ENST00000617271.4 ENST00000395446.5 ENST00000409834.5 ENST00000361849.7 ENST00000373957.7 ENST00000395430.5 ENST00000395433.5 ENST00000437009.5 |
PCDH15
|
protocadherin related 15 |
| chr22_-_30574511 | 0.15 |
ENST00000431313.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr6_-_39725387 | 0.15 |
ENST00000287152.12
|
KIF6
|
kinesin family member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.8 | 3.9 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.8 | 3.9 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.7 | 2.2 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.6 | 2.4 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.4 | 1.1 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 4.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.6 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.3 | 0.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 | 0.5 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.2 | 2.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.2 | 0.7 | GO:0042309 | octopamine biosynthetic process(GO:0006589) homoiothermy(GO:0042309) octopamine metabolic process(GO:0046333) |
| 0.2 | 3.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.6 | GO:0045175 | basal protein localization(GO:0045175) |
| 0.2 | 1.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.8 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.5 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.8 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.7 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 | 0.8 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.1 | 0.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 2.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.6 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.5 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 1.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 1.5 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.2 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 1.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.9 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.7 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.9 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 1.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 2.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 1.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 3.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 2.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.0 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 1.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 1.1 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.4 | 4.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 3.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 3.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 2.4 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 1.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 1.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 3.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.3 | 1.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 0.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 2.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 3.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.3 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.1 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.5 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 4.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 2.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 2.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0032181 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 3.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 4.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 2.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 5.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 3.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |