Project
avrg: Illumina Body Map 2 (GSE30611)
Navigation
Downloads
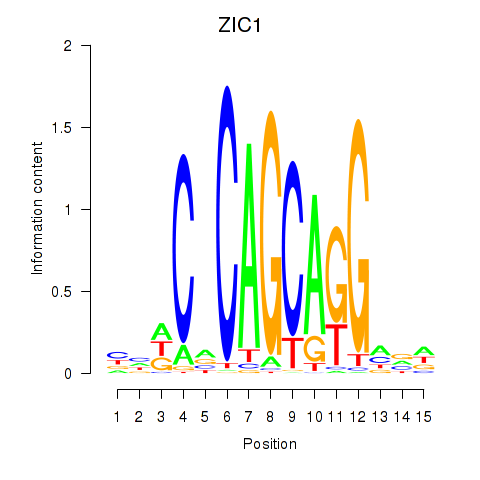
Results for ZIC1
Z-value: 1.58
Transcription factors associated with ZIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC1
|
ENSG00000152977.10 | ZIC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC1 | hg38_v1_chr3_+_147393718_147393895 | 0.01 | 9.7e-01 | Click! |
Activity profile of ZIC1 motif
Sorted Z-values of ZIC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_54588636 | 5.73 |
ENST00000257905.13
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A |
| chr11_+_116830529 | 5.10 |
ENST00000630701.1
|
APOC3
|
apolipoprotein C3 |
| chr1_+_65147514 | 3.75 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr5_+_151026966 | 3.60 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 |
| chr1_+_65147657 | 3.30 |
ENST00000546702.5
|
AK4
|
adenylate kinase 4 |
| chr19_+_44905785 | 3.30 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr14_-_64942720 | 2.99 |
ENST00000557049.1
ENST00000389614.6 |
GPX2
|
glutathione peroxidase 2 |
| chr14_-_64942783 | 2.75 |
ENST00000612794.1
|
GPX2
|
glutathione peroxidase 2 |
| chr3_-_12158901 | 2.65 |
ENST00000287814.5
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
| chr3_+_50155024 | 2.56 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chr13_-_28495079 | 2.53 |
ENST00000615840.4
ENST00000282397.9 ENST00000541932.5 ENST00000539099.1 ENST00000639477.1 |
FLT1
|
fms related receptor tyrosine kinase 1 |
| chr2_+_119431846 | 2.44 |
ENST00000306406.5
|
TMEM37
|
transmembrane protein 37 |
| chr12_-_54588516 | 2.27 |
ENST00000547431.5
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A |
| chr15_+_73684373 | 2.24 |
ENST00000558689.5
ENST00000560786.6 ENST00000318443.10 ENST00000561213.5 |
CD276
|
CD276 molecule |
| chr16_+_30985181 | 2.20 |
ENST00000262520.10
ENST00000297679.10 ENST00000562932.5 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_+_79120474 | 2.07 |
ENST00000233735.2
|
REG1A
|
regenerating family member 1 alpha |
| chr22_-_24226112 | 2.05 |
ENST00000425408.5
|
GGT5
|
gamma-glutamyltransferase 5 |
| chr4_+_110476133 | 2.04 |
ENST00000265162.10
|
ENPEP
|
glutamyl aminopeptidase |
| chr2_+_233760265 | 2.02 |
ENST00000305208.10
ENST00000360418.4 |
UGT1A1
|
UDP glucuronosyltransferase family 1 member A1 |
| chr22_+_43923755 | 2.02 |
ENST00000423180.2
ENST00000216180.8 |
PNPLA3
|
patatin like phospholipase domain containing 3 |
| chr11_-_2161158 | 1.99 |
ENST00000421783.1
ENST00000397262.5 ENST00000381330.5 ENST00000250971.7 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr10_-_33334625 | 1.95 |
ENST00000374875.5
ENST00000374822.8 ENST00000374867.7 |
NRP1
|
neuropilin 1 |
| chr2_-_1744442 | 1.93 |
ENST00000433670.5
ENST00000425171.1 ENST00000252804.9 |
PXDN
|
peroxidasin |
| chr15_+_73684204 | 1.88 |
ENST00000537340.6
ENST00000318424.9 |
CD276
|
CD276 molecule |
| chr10_-_33334898 | 1.88 |
ENST00000395995.5
|
NRP1
|
neuropilin 1 |
| chr4_-_185812209 | 1.73 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_-_42427379 | 1.70 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type |
| chr4_+_24795560 | 1.67 |
ENST00000382120.4
|
SOD3
|
superoxide dismutase 3 |
| chr8_-_119673368 | 1.66 |
ENST00000427067.6
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr19_+_41003946 | 1.62 |
ENST00000593831.1
|
CYP2B6
|
cytochrome P450 family 2 subfamily B member 6 |
| chr1_+_22636577 | 1.59 |
ENST00000374642.8
ENST00000438241.1 |
C1QA
|
complement C1q A chain |
| chr6_-_35688907 | 1.59 |
ENST00000539068.5
ENST00000357266.9 |
FKBP5
|
FKBP prolyl isomerase 5 |
| chr17_-_55732074 | 1.58 |
ENST00000575734.5
|
TMEM100
|
transmembrane protein 100 |
| chrX_-_130165699 | 1.54 |
ENST00000676328.1
ENST00000675857.1 ENST00000675427.1 ENST00000675092.1 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr11_+_67056805 | 1.53 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D |
| chrX_-_130165825 | 1.50 |
ENST00000675240.1
ENST00000319908.8 ENST00000674546.1 ENST00000287295.8 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr16_+_55479188 | 1.50 |
ENST00000219070.9
|
MMP2
|
matrix metallopeptidase 2 |
| chrX_-_130165664 | 1.42 |
ENST00000535724.6
ENST00000346424.6 ENST00000676436.1 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr9_-_109498251 | 1.41 |
ENST00000374541.4
ENST00000262539.7 |
PTPN3
|
protein tyrosine phosphatase non-receptor type 3 |
| chr17_-_44968263 | 1.40 |
ENST00000253407.4
|
C1QL1
|
complement C1q like 1 |
| chr1_-_59926724 | 1.37 |
ENST00000371204.4
|
CYP2J2
|
cytochrome P450 family 2 subfamily J member 2 |
| chr1_+_16043736 | 1.35 |
ENST00000619181.4
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chrX_-_130165873 | 1.34 |
ENST00000676229.1
|
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr8_-_63038788 | 1.33 |
ENST00000518113.2
ENST00000260118.7 ENST00000677482.1 |
GGH
|
gamma-glutamyl hydrolase |
| chr12_+_109154661 | 1.24 |
ENST00000544726.2
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr10_-_33335074 | 1.24 |
ENST00000432372.6
|
NRP1
|
neuropilin 1 |
| chr1_+_16043776 | 1.23 |
ENST00000375679.9
|
CLCNKB
|
chloride voltage-gated channel Kb |
| chr6_-_35688935 | 1.22 |
ENST00000542713.1
|
FKBP5
|
FKBP prolyl isomerase 5 |
| chr1_+_16021871 | 1.20 |
ENST00000375692.5
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr1_+_16022030 | 1.14 |
ENST00000331433.5
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr9_-_35103178 | 1.12 |
ENST00000452248.6
ENST00000619795.4 |
STOML2
|
stomatin like 2 |
| chr17_+_41265339 | 1.11 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr4_+_6269831 | 1.11 |
ENST00000503569.5
ENST00000673991.1 ENST00000682275.1 ENST00000226760.5 |
WFS1
|
wolframin ER transmembrane glycoprotein |
| chr11_+_67056875 | 1.09 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr4_-_128286875 | 1.09 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr21_-_38498415 | 1.05 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr19_+_44809089 | 1.05 |
ENST00000270233.12
ENST00000591520.6 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr9_-_35103108 | 1.04 |
ENST00000356493.10
|
STOML2
|
stomatin like 2 |
| chr1_+_16022004 | 1.01 |
ENST00000439316.6
|
CLCNKA
|
chloride voltage-gated channel Ka |
| chr4_-_128286787 | 1.00 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr17_-_69060906 | 0.97 |
ENST00000495634.5
ENST00000453985.6 ENST00000340001.9 ENST00000585714.1 |
ABCA9
|
ATP binding cassette subfamily A member 9 |
| chr3_+_149129610 | 0.96 |
ENST00000460120.5
ENST00000296051.7 |
HPS3
|
HPS3 biogenesis of lysosomal organelles complex 2 subunit 1 |
| chr19_+_46303599 | 0.96 |
ENST00000300862.7
|
HIF3A
|
hypoxia inducible factor 3 subunit alpha |
| chr3_-_73433904 | 0.95 |
ENST00000479530.5
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr1_+_61404076 | 0.92 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr3_-_58577367 | 0.91 |
ENST00000464064.5
ENST00000360997.7 |
FAM107A
|
family with sequence similarity 107 member A |
| chr9_+_121567057 | 0.91 |
ENST00000394340.7
ENST00000436835.5 ENST00000259371.6 |
DAB2IP
|
DAB2 interacting protein |
| chr12_-_106084023 | 0.89 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family kinase 1 |
| chr3_+_51977833 | 0.87 |
ENST00000637978.1
|
ABHD14A-ACY1
|
ABHD14A-ACY1 readthrough |
| chr2_+_6978624 | 0.87 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr2_-_75518697 | 0.86 |
ENST00000452003.1
|
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chrX_+_46912412 | 0.86 |
ENST00000614628.5
|
JADE3
|
jade family PHD finger 3 |
| chr7_-_150800533 | 0.86 |
ENST00000434545.5
|
TMEM176B
|
transmembrane protein 176B |
| chr1_-_153616289 | 0.83 |
ENST00000368701.5
ENST00000344616.4 |
S100A14
|
S100 calcium binding protein A14 |
| chr6_-_116765709 | 0.82 |
ENST00000368557.6
|
FAM162B
|
family with sequence similarity 162 member B |
| chr13_-_44474296 | 0.81 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1 |
| chr19_+_44809053 | 0.81 |
ENST00000611077.5
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr5_-_150137403 | 0.81 |
ENST00000517957.1
|
PDGFRB
|
platelet derived growth factor receptor beta |
| chr10_-_69416323 | 0.80 |
ENST00000619173.4
|
TACR2
|
tachykinin receptor 2 |
| chr14_+_38207893 | 0.80 |
ENST00000267377.3
|
SSTR1
|
somatostatin receptor 1 |
| chr13_-_44474250 | 0.80 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr19_-_55117336 | 0.77 |
ENST00000592993.1
|
PPP1R12C
|
protein phosphatase 1 regulatory subunit 12C |
| chr19_-_3028356 | 0.72 |
ENST00000586422.5
|
TLE2
|
TLE family member 2, transcriptional corepressor |
| chr1_-_43367956 | 0.71 |
ENST00000372458.8
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chrX_+_46912276 | 0.71 |
ENST00000424392.5
ENST00000611250.4 |
JADE3
|
jade family PHD finger 3 |
| chr15_+_84980440 | 0.70 |
ENST00000310298.8
ENST00000557957.5 |
PDE8A
|
phosphodiesterase 8A |
| chr1_+_61952036 | 0.67 |
ENST00000646453.1
ENST00000635137.1 |
PATJ
|
PATJ crumbs cell polarity complex component |
| chr3_+_42850966 | 0.66 |
ENST00000494619.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr14_-_68979436 | 0.66 |
ENST00000193403.10
|
ACTN1
|
actinin alpha 1 |
| chr3_-_86991135 | 0.66 |
ENST00000398399.7
|
VGLL3
|
vestigial like family member 3 |
| chr12_+_77324608 | 0.65 |
ENST00000550042.2
|
NAV3
|
neuron navigator 3 |
| chr19_-_16660104 | 0.63 |
ENST00000593459.5
ENST00000627144.2 ENST00000358726.6 ENST00000487416.7 ENST00000597711.5 |
ENSG00000268790.5
SMIM7
|
novel protein small integral membrane protein 7 |
| chr19_-_14475307 | 0.61 |
ENST00000292513.4
|
PTGER1
|
prostaglandin E receptor 1 |
| chr1_-_43368039 | 0.61 |
ENST00000413844.3
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr8_+_84184875 | 0.61 |
ENST00000517638.5
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein like |
| chr17_-_58531941 | 0.60 |
ENST00000581607.1
ENST00000317256.10 ENST00000426861.5 ENST00000580809.5 ENST00000577729.5 ENST00000583291.1 |
SEPTIN4
|
septin 4 |
| chr3_-_50292404 | 0.59 |
ENST00000417626.8
|
IFRD2
|
interferon related developmental regulator 2 |
| chr12_-_102478539 | 0.59 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr14_-_68979274 | 0.58 |
ENST00000394419.9
|
ACTN1
|
actinin alpha 1 |
| chr1_+_99850485 | 0.58 |
ENST00000370165.7
ENST00000370163.7 ENST00000294724.8 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr17_-_979726 | 0.57 |
ENST00000336868.8
|
NXN
|
nucleoredoxin |
| chr14_-_68979314 | 0.57 |
ENST00000684713.1
ENST00000683198.1 ENST00000684598.1 ENST00000682331.1 ENST00000682291.1 ENST00000683342.1 |
ACTN1
|
actinin alpha 1 |
| chrX_+_46912629 | 0.57 |
ENST00000455411.1
|
JADE3
|
jade family PHD finger 3 |
| chr2_-_68319398 | 0.56 |
ENST00000409559.7
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr1_-_43367689 | 0.55 |
ENST00000621943.4
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr4_-_185811965 | 0.55 |
ENST00000419063.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_123754518 | 0.54 |
ENST00000327930.3
|
OR6X1
|
olfactory receptor family 6 subfamily X member 1 |
| chr7_-_108240049 | 0.54 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr15_+_73683938 | 0.54 |
ENST00000567189.5
|
CD276
|
CD276 molecule |
| chr22_-_30387078 | 0.53 |
ENST00000215798.10
|
RNF215
|
ring finger protein 215 |
| chr16_-_31088963 | 0.53 |
ENST00000280606.6
|
PRSS53
|
serine protease 53 |
| chr3_-_86991109 | 0.53 |
ENST00000383698.3
|
VGLL3
|
vestigial like family member 3 |
| chr1_+_23793649 | 0.52 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase 2 |
| chr14_+_22871732 | 0.51 |
ENST00000359591.9
|
LRP10
|
LDL receptor related protein 10 |
| chr1_+_46203321 | 0.51 |
ENST00000371980.4
|
LURAP1
|
leucine rich adaptor protein 1 |
| chr4_+_6269869 | 0.51 |
ENST00000506362.2
|
WFS1
|
wolframin ER transmembrane glycoprotein |
| chrX_+_69616067 | 0.51 |
ENST00000338901.4
ENST00000525810.5 ENST00000527388.5 ENST00000374553.6 ENST00000374552.9 ENST00000524573.5 |
EDA
|
ectodysplasin A |
| chr5_-_127073467 | 0.49 |
ENST00000607731.5
ENST00000509733.7 ENST00000296662.10 ENST00000535381.6 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr7_-_73842508 | 0.49 |
ENST00000297873.9
|
METTL27
|
methyltransferase like 27 |
| chr14_-_21634940 | 0.48 |
ENST00000542433.1
|
OR10G2
|
olfactory receptor family 10 subfamily G member 2 |
| chr19_-_35501878 | 0.48 |
ENST00000593342.5
ENST00000601650.1 ENST00000408915.6 |
DMKN
|
dermokine |
| chr20_+_56629296 | 0.47 |
ENST00000201031.3
|
TFAP2C
|
transcription factor AP-2 gamma |
| chr22_-_30557586 | 0.47 |
ENST00000338911.6
ENST00000453479.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr15_+_59372685 | 0.46 |
ENST00000558348.5
|
FAM81A
|
family with sequence similarity 81 member A |
| chr6_-_168075831 | 0.46 |
ENST00000440994.6
|
FRMD1
|
FERM domain containing 1 |
| chr2_+_241813947 | 0.45 |
ENST00000428592.1
|
NEU4
|
neuraminidase 4 |
| chr1_+_18631513 | 0.45 |
ENST00000400661.3
|
PAX7
|
paired box 7 |
| chr1_+_99850348 | 0.44 |
ENST00000361915.8
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr17_-_58544315 | 0.43 |
ENST00000671766.1
ENST00000672673.2 ENST00000321691.3 |
SEPTIN4
|
septin 4 |
| chr6_+_31762996 | 0.42 |
ENST00000415669.3
ENST00000425424.4 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr16_+_28846637 | 0.41 |
ENST00000567536.5
|
SH2B1
|
SH2B adaptor protein 1 |
| chr14_-_68979251 | 0.41 |
ENST00000438964.6
ENST00000679147.1 |
ACTN1
|
actinin alpha 1 |
| chr12_+_56996151 | 0.40 |
ENST00000556850.1
|
GPR182
|
G protein-coupled receptor 182 |
| chr1_+_18631006 | 0.40 |
ENST00000375375.7
|
PAX7
|
paired box 7 |
| chr1_-_111449209 | 0.40 |
ENST00000235090.10
|
WDR77
|
WD repeat domain 77 |
| chr14_-_68979076 | 0.40 |
ENST00000538545.6
ENST00000684639.1 |
ACTN1
|
actinin alpha 1 |
| chr15_+_73684435 | 0.39 |
ENST00000563584.5
ENST00000561416.5 |
CD276
|
CD276 molecule |
| chr12_-_58920465 | 0.39 |
ENST00000320743.8
|
LRIG3
|
leucine rich repeats and immunoglobulin like domains 3 |
| chr9_-_122629573 | 0.39 |
ENST00000623530.1
|
OR1B1
|
olfactory receptor family 1 subfamily B member 1 |
| chr1_+_111449442 | 0.39 |
ENST00000369722.8
ENST00000483994.1 |
ATP5PB
|
ATP synthase peripheral stalk-membrane subunit b |
| chr11_-_59845496 | 0.39 |
ENST00000257248.3
|
CBLIF
|
cobalamin binding intrinsic factor |
| chr17_-_41168219 | 0.38 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chrX_+_3006546 | 0.38 |
ENST00000381130.3
|
ARSH
|
arylsulfatase family member H |
| chr1_-_89175997 | 0.38 |
ENST00000294671.3
ENST00000650452.1 |
GBP7
|
guanylate binding protein 7 |
| chrX_-_47574738 | 0.38 |
ENST00000640721.1
|
SYN1
|
synapsin I |
| chr16_+_28846596 | 0.37 |
ENST00000563591.5
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_94121105 | 0.36 |
ENST00000649773.1
ENST00000370225.4 |
ABCA4
|
ATP binding cassette subfamily A member 4 |
| chr6_-_168319762 | 0.36 |
ENST00000366795.4
|
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr3_+_4303602 | 0.36 |
ENST00000358950.4
|
SETMAR
|
SET domain and mariner transposase fusion gene |
| chr11_+_89710650 | 0.35 |
ENST00000534392.4
|
TRIM77
|
tripartite motif containing 77 |
| chr11_+_75815180 | 0.35 |
ENST00000356136.8
|
UVRAG
|
UV radiation resistance associated |
| chr1_+_161118083 | 0.35 |
ENST00000368009.7
ENST00000368007.8 ENST00000392190.9 ENST00000368008.5 |
NIT1
|
nitrilase 1 |
| chr21_+_30396030 | 0.34 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr11_+_58179431 | 0.33 |
ENST00000612174.1
|
OR9Q1
|
olfactory receptor family 9 subfamily Q member 1 |
| chr19_-_58098203 | 0.33 |
ENST00000600845.1
ENST00000240727.10 ENST00000600897.5 ENST00000421612.6 ENST00000601063.1 ENST00000601144.6 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr2_-_96868575 | 0.33 |
ENST00000442264.5
|
SEMA4C
|
semaphorin 4C |
| chr1_+_61952283 | 0.33 |
ENST00000307297.8
|
PATJ
|
PATJ crumbs cell polarity complex component |
| chr17_-_41027198 | 0.32 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr6_-_168319691 | 0.31 |
ENST00000610183.1
ENST00000607983.1 |
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr6_+_132570322 | 0.31 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr8_-_81841958 | 0.31 |
ENST00000519817.5
ENST00000521773.5 ENST00000523757.5 ENST00000345957.9 |
SNX16
|
sorting nexin 16 |
| chr12_+_5432101 | 0.31 |
ENST00000423158.4
|
NTF3
|
neurotrophin 3 |
| chr14_-_23548437 | 0.30 |
ENST00000555334.1
|
ZFHX2
|
zinc finger homeobox 2 |
| chr3_-_45796518 | 0.30 |
ENST00000413781.1
ENST00000358525.9 |
SLC6A20
|
solute carrier family 6 member 20 |
| chr10_+_49610297 | 0.29 |
ENST00000374115.5
|
SLC18A3
|
solute carrier family 18 member A3 |
| chr13_+_101452569 | 0.29 |
ENST00000618057.4
|
ITGBL1
|
integrin subunit beta like 1 |
| chr14_+_57268940 | 0.28 |
ENST00000556995.1
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr11_-_2171805 | 0.28 |
ENST00000381178.5
ENST00000381175.5 ENST00000333684.9 ENST00000352909.8 |
TH
|
tyrosine hydroxylase |
| chr9_-_16870702 | 0.28 |
ENST00000380667.6
ENST00000545497.5 ENST00000486514.5 |
BNC2
|
basonuclin 2 |
| chr1_+_171600621 | 0.27 |
ENST00000636697.1
|
MYOCOS
|
myocilin opposite strand |
| chr3_-_45796467 | 0.27 |
ENST00000353278.8
ENST00000456124.6 |
SLC6A20
|
solute carrier family 6 member 20 |
| chr4_-_4226929 | 0.25 |
ENST00000296358.5
|
OTOP1
|
otopetrin 1 |
| chr6_+_89562620 | 0.25 |
ENST00000522779.5
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr1_-_211133945 | 0.24 |
ENST00000640044.1
ENST00000640566.1 |
KCNH1
|
potassium voltage-gated channel subfamily H member 1 |
| chr19_-_6433754 | 0.23 |
ENST00000321510.7
|
SLC25A41
|
solute carrier family 25 member 41 |
| chr22_-_30591850 | 0.23 |
ENST00000335214.8
ENST00000406208.7 ENST00000402284.7 ENST00000354694.12 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr11_+_89710299 | 0.23 |
ENST00000398290.7
|
TRIM77
|
tripartite motif containing 77 |
| chr8_-_81842192 | 0.22 |
ENST00000353788.8
ENST00000520618.5 ENST00000518183.5 ENST00000396330.6 ENST00000519119.5 |
SNX16
|
sorting nexin 16 |
| chr21_+_44697427 | 0.21 |
ENST00000618832.1
|
KRTAP10-12
|
keratin associated protein 10-12 |
| chr17_+_38428456 | 0.21 |
ENST00000622683.5
ENST00000620417.4 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr11_+_15073562 | 0.21 |
ENST00000533448.1
ENST00000324229.11 |
CALCB
|
calcitonin related polypeptide beta |
| chr17_-_41065879 | 0.20 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr4_-_40515967 | 0.19 |
ENST00000381795.10
|
RBM47
|
RNA binding motif protein 47 |
| chr4_+_26576670 | 0.19 |
ENST00000512840.5
|
TBC1D19
|
TBC1 domain family member 19 |
| chr17_-_5468951 | 0.19 |
ENST00000225296.8
|
DHX33
|
DEAH-box helicase 33 |
| chr20_+_33031648 | 0.19 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B member 6 |
| chr21_+_30440275 | 0.18 |
ENST00000334067.4
|
KRTAP15-1
|
keratin associated protein 15-1 |
| chr6_-_168319744 | 0.17 |
ENST00000366796.7
|
DACT2
|
dishevelled binding antagonist of beta catenin 2 |
| chr6_+_89562766 | 0.17 |
ENST00000485637.5
ENST00000522705.5 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr21_-_44540195 | 0.17 |
ENST00000400375.1
|
KRTAP10-1
|
keratin associated protein 10-1 |
| chr14_+_57269007 | 0.16 |
ENST00000431972.6
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr17_+_45999250 | 0.16 |
ENST00000537309.1
|
STH
|
saitohin |
| chr4_+_94995919 | 0.16 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr7_-_103149056 | 0.16 |
ENST00000465647.6
ENST00000418294.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr22_+_29306582 | 0.15 |
ENST00000616432.4
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest specific 2 like 1 |
| chr3_-_79767987 | 0.15 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1 |
| chr11_-_559377 | 0.15 |
ENST00000486629.1
|
LMNTD2
|
lamin tail domain containing 2 |
| chr18_+_63752935 | 0.15 |
ENST00000425392.5
ENST00000336429.6 |
SERPINB7
|
serpin family B member 7 |
| chr1_-_244730962 | 0.14 |
ENST00000640271.2
|
ENSG00000284188.2
|
novel protein |
| chr14_+_57268963 | 0.14 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 1.4 | 8.4 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 1.3 | 5.1 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.8 | 2.5 | GO:0036323 | vascular endothelial growth factor receptor-1 signaling pathway(GO:0036323) |
| 0.7 | 2.0 | GO:0060266 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.5 | 7.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.5 | 3.6 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.5 | 2.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.4 | 2.2 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.4 | 1.6 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.4 | 4.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 2.6 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.3 | 2.0 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.3 | 1.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 0.8 | GO:0014813 | skeletal muscle satellite cell commitment(GO:0014813) |
| 0.3 | 5.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 0.3 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.3 | 0.8 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.3 | 1.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 1.4 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 2.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 0.8 | GO:1900108 | inner medullary collecting duct development(GO:0072061) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.8 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 0.8 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 1.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.9 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.0 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 2.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 3.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.6 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.9 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 2.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 1.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.4 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.6 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 2.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 5.9 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.9 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 9.0 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.1 | 0.9 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 9.9 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.3 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 2.9 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.8 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.6 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.5 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0021825 | substrate-dependent cerebral cortex tangential migration(GO:0021825) |
| 0.0 | 1.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.7 | GO:1903206 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 3.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 5.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 1.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.3 | 2.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.2 | 1.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 8.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 6.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 2.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 6.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 4.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 1.0 | 5.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.8 | 2.5 | GO:0036326 | VEGF-A-activated receptor activity(GO:0036326) VEGF-B-activated receptor activity(GO:0036327) placental growth factor-activated receptor activity(GO:0036332) |
| 0.7 | 3.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.6 | 1.9 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.6 | 1.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.5 | 5.9 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.5 | 8.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 5.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 1.9 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 1.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.3 | 0.8 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.3 | 9.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 2.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 2.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 5.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 1.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 3.0 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.9 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 3.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 2.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 2.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 2.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 2.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 2.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.3 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 2.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.8 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.0 | 2.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 5.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 5.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 8.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |