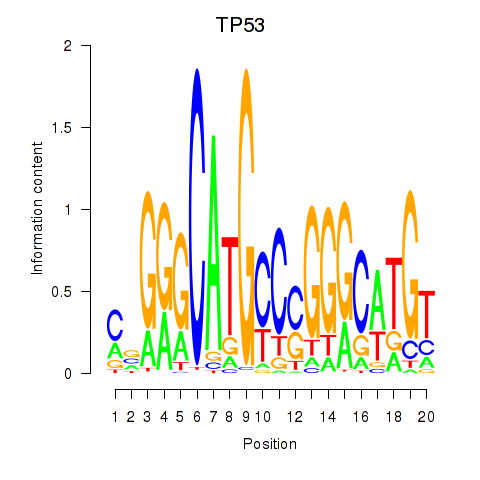
|
chr14_-_106235582
Show fit
|
5.53 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21
|
|
chr8_+_23102913
Show fit
|
5.20 |
ENST00000356864.4
|
TNFRSF10C
|
TNF receptor superfamily member 10c
|
|
chr11_-_47378527
Show fit
|
5.15 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr11_-_47378391
Show fit
|
4.84 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr11_-_47378494
Show fit
|
4.84 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr11_-_47378479
Show fit
|
4.65 |
ENST00000533968.1
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr11_+_65879791
Show fit
|
4.12 |
ENST00000528419.6
ENST00000307886.8
ENST00000526034.2
ENST00000679584.1
ENST00000680443.1
ENST00000680670.1
|
CTSW
|
cathepsin W
|
|
chr18_+_59899988
Show fit
|
3.96 |
ENST00000316660.7
ENST00000269518.9
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr8_-_23225061
Show fit
|
3.94 |
ENST00000613472.1
ENST00000221132.8
|
TNFRSF10A
|
TNF receptor superfamily member 10a
|
|
chr19_+_3136026
Show fit
|
3.36 |
ENST00000262958.4
|
GNA15
|
G protein subunit alpha 15
|
|
chr8_-_23164020
Show fit
|
3.26 |
ENST00000312584.4
|
TNFRSF10D
|
TNF receptor superfamily member 10d
|
|
chr2_-_234497035
Show fit
|
3.23 |
ENST00000390645.2
ENST00000339728.6
|
ARL4C
|
ADP ribosylation factor like GTPase 4C
|
|
chr8_-_10730498
Show fit
|
3.21 |
ENST00000304501.2
|
SOX7
|
SRY-box transcription factor 7
|
|
chr20_-_1393045
Show fit
|
3.07 |
ENST00000400137.9
ENST00000381715.4
|
FKBP1A
|
FKBP prolyl isomerase 1A
|
|
chr7_+_99374722
Show fit
|
3.01 |
ENST00000418347.6
ENST00000429246.6
ENST00000417330.6
ENST00000431816.6
ENST00000458033.6
ENST00000451682.5
ENST00000427217.6
ENST00000646101.2
|
ARPC1B
|
actin related protein 2/3 complex subunit 1B
|
|
chr1_-_150765735
Show fit
|
3.00 |
ENST00000679898.1
ENST00000448301.7
ENST00000680664.1
ENST00000679512.1
ENST00000368985.8
ENST00000679582.1
|
CTSS
|
cathepsin S
|
|
chr19_+_827823
Show fit
|
2.97 |
ENST00000233997.4
|
AZU1
|
azurocidin 1
|
|
chr1_-_37947010
Show fit
|
2.90 |
ENST00000458109.6
ENST00000373024.8
ENST00000373023.6
|
INPP5B
|
inositol polyphosphate-5-phosphatase B
|
|
chr9_+_36136416
Show fit
|
2.79 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2
|
|
chr20_-_1393074
Show fit
|
2.78 |
ENST00000614856.2
ENST00000678408.1
ENST00000618612.5
ENST00000439640.5
ENST00000381719.8
ENST00000677533.1
|
FKBP1A
|
FKBP prolyl isomerase 1A
|
|
chr5_-_132490750
Show fit
|
2.57 |
ENST00000437654.6
ENST00000245414.9
ENST00000680139.1
ENST00000680352.1
ENST00000679440.1
ENST00000680903.1
|
IRF1
|
interferon regulatory factor 1
|
|
chr6_+_36130484
Show fit
|
2.46 |
ENST00000373766.9
ENST00000211287.9
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr1_+_161524539
Show fit
|
2.44 |
ENST00000309758.6
|
HSPA6
|
heat shock protein family A (Hsp70) member 6
|
|
chr11_-_65859282
Show fit
|
2.43 |
ENST00000526975.1
ENST00000531413.5
ENST00000525451.6
|
CFL1
|
cofilin 1
|
|
chr7_+_99374675
Show fit
|
2.42 |
ENST00000645391.1
ENST00000455009.6
|
ARPC1B
|
actin related protein 2/3 complex subunit 1B
|
|
chr19_+_48954850
Show fit
|
2.40 |
ENST00000345358.12
ENST00000539787.2
ENST00000415969.6
ENST00000354470.7
ENST00000506183.5
ENST00000391871.4
ENST00000293288.12
|
BAX
|
BCL2 associated X, apoptosis regulator
|
|
chr1_+_27879638
Show fit
|
2.38 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2
|
|
chr11_-_44950839
Show fit
|
2.38 |
ENST00000395648.7
ENST00000531928.6
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr10_+_88990736
Show fit
|
2.35 |
ENST00000357339.6
ENST00000652046.1
ENST00000355279.2
|
FAS
|
Fas cell surface death receptor
|
|
chr19_-_6481769
Show fit
|
2.33 |
ENST00000381480.7
ENST00000543576.5
ENST00000590173.5
|
DENND1C
|
DENN domain containing 1C
|
|
chr1_-_160862700
Show fit
|
2.32 |
ENST00000322302.7
ENST00000368033.7
|
CD244
|
CD244 molecule
|
|
chr17_+_14301069
Show fit
|
2.31 |
ENST00000360954.3
|
HS3ST3B1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1
|
|
chr1_-_150765785
Show fit
|
2.30 |
ENST00000680311.1
ENST00000681728.1
ENST00000680288.1
|
CTSS
|
cathepsin S
|
|
chr8_-_23069012
Show fit
|
2.28 |
ENST00000347739.3
ENST00000276431.9
|
TNFRSF10B
|
TNF receptor superfamily member 10b
|
|
chr10_-_103452356
Show fit
|
2.24 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr1_-_160862880
Show fit
|
2.24 |
ENST00000368034.9
|
CD244
|
CD244 molecule
|
|
chr1_-_54018167
Show fit
|
2.23 |
ENST00000371362.7
ENST00000371360.2
ENST00000545928.5
ENST00000420619.5
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr16_+_527698
Show fit
|
2.23 |
ENST00000219611.7
ENST00000562370.5
ENST00000568988.5
|
CAPN15
|
calpain 15
|
|
chr19_-_42217667
Show fit
|
2.19 |
ENST00000336034.8
ENST00000596251.6
ENST00000598200.1
ENST00000598727.5
|
DEDD2
|
death effector domain containing 2
|
|
chr11_+_67403887
Show fit
|
2.18 |
ENST00000526387.5
|
TBC1D10C
|
TBC1 domain family member 10C
|
|
chr6_-_30744537
Show fit
|
2.16 |
ENST00000259874.6
ENST00000376377.2
|
IER3
|
immediate early response 3
|
|
chr11_-_73982830
Show fit
|
2.14 |
ENST00000536983.5
ENST00000663595.2
ENST00000310473.9
|
UCP2
|
uncoupling protein 2
|
|
chr19_+_41860236
Show fit
|
2.14 |
ENST00000601492.5
ENST00000600467.6
ENST00000598742.6
ENST00000221975.6
ENST00000598261.2
|
RPS19
|
ribosomal protein S19
|
|
chr11_+_5691004
Show fit
|
2.12 |
ENST00000414641.5
|
TRIM22
|
tripartite motif containing 22
|
|
chr1_+_17308194
Show fit
|
2.11 |
ENST00000375453.5
ENST00000375448.4
|
PADI4
|
peptidyl arginine deiminase 4
|
|
chrX_-_154354889
Show fit
|
2.09 |
ENST00000438732.2
|
FLNA
|
filamin A
|
|
chr19_-_47232649
Show fit
|
2.06 |
ENST00000449228.5
ENST00000300880.11
ENST00000341983.8
|
BBC3
|
BCL2 binding component 3
|
|
chr6_-_132714045
Show fit
|
2.05 |
ENST00000367928.5
|
VNN1
|
vanin 1
|
|
chr1_+_9997220
Show fit
|
2.02 |
ENST00000294435.8
|
RBP7
|
retinol binding protein 7
|
|
chr7_+_48088596
Show fit
|
1.97 |
ENST00000416681.5
ENST00000331803.8
ENST00000432131.5
|
UPP1
|
uridine phosphorylase 1
|
|
chrX_+_100644183
Show fit
|
1.97 |
ENST00000640889.1
ENST00000373004.5
|
SRPX2
|
sushi repeat containing protein X-linked 2
|
|
chr19_+_56141317
Show fit
|
1.86 |
ENST00000592949.5
|
ZNF444
|
zinc finger protein 444
|
|
chr19_-_51646800
Show fit
|
1.83 |
ENST00000599649.5
ENST00000429354.3
ENST00000360844.6
|
SIGLEC5
SIGLEC14
|
sialic acid binding Ig like lectin 5
sialic acid binding Ig like lectin 14
|
|
chr10_-_103452384
Show fit
|
1.77 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr20_+_35172046
Show fit
|
1.76 |
ENST00000216968.5
|
PROCR
|
protein C receptor
|
|
chr3_-_39281663
Show fit
|
1.70 |
ENST00000358309.3
|
CX3CR1
|
C-X3-C motif chemokine receptor 1
|
|
chr17_-_63446168
Show fit
|
1.69 |
ENST00000584031.5
ENST00000392976.5
|
CYB561
|
cytochrome b561
|
|
chr3_-_52445085
Show fit
|
1.69 |
ENST00000475739.1
ENST00000231721.7
|
SEMA3G
|
semaphorin 3G
|
|
chr17_-_63446260
Show fit
|
1.68 |
ENST00000448884.6
ENST00000582297.5
ENST00000360793.8
ENST00000582034.5
ENST00000578072.1
|
CYB561
|
cytochrome b561
|
|
chr12_-_14567714
Show fit
|
1.68 |
ENST00000240617.10
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr19_+_56141277
Show fit
|
1.67 |
ENST00000337080.8
ENST00000586123.5
|
ZNF444
|
zinc finger protein 444
|
|
chr1_+_26826682
Show fit
|
1.66 |
ENST00000374142.9
|
ZDHHC18
|
zinc finger DHHC-type palmitoyltransferase 18
|
|
chr4_+_25648011
Show fit
|
1.65 |
ENST00000645788.1
|
SLC34A2
|
solute carrier family 34 member 2
|
|
chr1_+_11736120
Show fit
|
1.63 |
ENST00000400895.6
ENST00000314340.10
ENST00000376629.8
ENST00000376627.6
ENST00000452018.6
ENST00000510878.1
|
AGTRAP
|
angiotensin II receptor associated protein
|
|
chr15_-_64162965
Show fit
|
1.62 |
ENST00000300026.4
ENST00000681397.1
ENST00000681658.1
|
PPIB
|
peptidylprolyl isomerase B
|
|
chr3_+_52495330
Show fit
|
1.62 |
ENST00000321725.10
|
STAB1
|
stabilin 1
|
|
chr3_-_12759224
Show fit
|
1.59 |
ENST00000314124.12
ENST00000435218.6
ENST00000435575.5
|
TMEM40
|
transmembrane protein 40
|
|
chr9_-_133121148
Show fit
|
1.59 |
ENST00000372047.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr1_-_209652329
Show fit
|
1.58 |
ENST00000367030.7
ENST00000356082.9
|
LAMB3
|
laminin subunit beta 3
|
|
chr19_-_6591103
Show fit
|
1.57 |
ENST00000423145.7
ENST00000245903.4
|
CD70
|
CD70 molecule
|
|
chr7_-_38349972
Show fit
|
1.55 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr22_-_39319595
Show fit
|
1.54 |
ENST00000427905.5
ENST00000216146.9
ENST00000402527.5
|
RPL3
|
ribosomal protein L3
|
|
chr1_-_156020789
Show fit
|
1.51 |
ENST00000531917.5
ENST00000480567.5
ENST00000526212.2
ENST00000529008.5
ENST00000496742.5
ENST00000295702.9
|
SSR2
|
signal sequence receptor subunit 2
|
|
chr16_-_14694308
Show fit
|
1.49 |
ENST00000438167.8
|
PLA2G10
|
phospholipase A2 group X
|
|
chr1_+_11736068
Show fit
|
1.48 |
ENST00000376637.7
|
AGTRAP
|
angiotensin II receptor associated protein
|
|
chr9_-_127755030
Show fit
|
1.48 |
ENST00000414380.1
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr4_-_10116724
Show fit
|
1.45 |
ENST00000502702.5
|
WDR1
|
WD repeat domain 1
|
|
chr14_+_92513766
Show fit
|
1.44 |
ENST00000216487.12
ENST00000620541.4
ENST00000557762.1
|
RIN3
|
Ras and Rab interactor 3
|
|
chr19_-_54364983
Show fit
|
1.43 |
ENST00000434277.6
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1
|
|
chr11_-_68844252
Show fit
|
1.43 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A
|
|
chr7_+_101127095
Show fit
|
1.42 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1
|
|
chr20_+_62952674
Show fit
|
1.42 |
ENST00000370349.7
ENST00000370351.9
|
SLC17A9
|
solute carrier family 17 member 9
|
|
chr10_+_46375721
Show fit
|
1.41 |
ENST00000616785.1
ENST00000611655.1
ENST00000619162.5
|
ENSG00000285402.1
ANXA8L1
|
novel transcript
annexin A8 like 1
|
|
chr17_+_27294076
Show fit
|
1.41 |
ENST00000581440.5
ENST00000583742.1
ENST00000579733.5
ENST00000583193.5
ENST00000581185.5
ENST00000427287.6
ENST00000262394.7
ENST00000348811.6
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr17_+_76376581
Show fit
|
1.40 |
ENST00000591651.5
ENST00000545180.5
|
SPHK1
|
sphingosine kinase 1
|
|
chr10_-_47484081
Show fit
|
1.39 |
ENST00000583448.2
ENST00000583874.5
ENST00000585281.6
|
ANXA8
|
annexin A8
|
|
chr1_-_3611470
Show fit
|
1.38 |
ENST00000356575.9
|
MEGF6
|
multiple EGF like domains 6
|
|
chr1_-_154968874
Show fit
|
1.36 |
ENST00000444179.5
ENST00000414115.5
|
SHC1
|
SHC adaptor protein 1
|
|
chr10_+_46375619
Show fit
|
1.33 |
ENST00000584982.7
ENST00000613703.4
|
ANXA8L1
|
annexin A8 like 1
|
|
chr19_+_19385815
Show fit
|
1.29 |
ENST00000494516.6
ENST00000360315.7
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr19_+_3585473
Show fit
|
1.28 |
ENST00000644452.3
ENST00000644946.1
|
GIPC3
|
GIPC PDZ domain containing family member 3
|
|
chr10_-_47484133
Show fit
|
1.27 |
ENST00000583911.5
ENST00000611843.4
|
ANXA8
|
annexin A8
|
|
chr10_-_71851313
Show fit
|
1.27 |
ENST00000394934.4
ENST00000610929.3
|
PSAP
|
prosaposin
|
|
chr2_+_112645930
Show fit
|
1.25 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1
|
|
chr20_+_44966478
Show fit
|
1.25 |
ENST00000499879.6
ENST00000372806.8
ENST00000372801.5
|
STK4
|
serine/threonine kinase 4
|
|
chr9_-_133121228
Show fit
|
1.24 |
ENST00000372050.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr12_-_6342066
Show fit
|
1.23 |
ENST00000162749.7
ENST00000440083.6
|
TNFRSF1A
|
TNF receptor superfamily member 1A
|
|
chr19_+_926001
Show fit
|
1.23 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A
|
|
chr12_-_6342020
Show fit
|
1.22 |
ENST00000540022.5
ENST00000536194.1
|
TNFRSF1A
|
TNF receptor superfamily member 1A
|
|
chr10_-_125158704
Show fit
|
1.19 |
ENST00000531469.5
|
CTBP2
|
C-terminal binding protein 2
|
|
chr11_-_75306688
Show fit
|
1.19 |
ENST00000532525.1
|
ARRB1
|
arrestin beta 1
|
|
chr7_+_43583091
Show fit
|
1.19 |
ENST00000319357.6
|
STK17A
|
serine/threonine kinase 17a
|
|
chr16_+_8797716
Show fit
|
1.18 |
ENST00000682008.1
|
PMM2
|
phosphomannomutase 2
|
|
chr16_+_57372525
Show fit
|
1.18 |
ENST00000564948.1
|
CX3CL1
|
C-X3-C motif chemokine ligand 1
|
|
chr17_+_27456393
Show fit
|
1.18 |
ENST00000644974.1
|
KSR1
|
kinase suppressor of ras 1
|
|
chr12_-_6341848
Show fit
|
1.17 |
ENST00000366159.8
ENST00000539372.5
|
TNFRSF1A
|
TNF receptor superfamily member 1A
|
|
chr2_+_95025700
Show fit
|
1.14 |
ENST00000309988.9
ENST00000353004.7
ENST00000354078.7
ENST00000349807.3
|
MAL
|
mal, T cell differentiation protein
|
|
chr6_+_36130586
Show fit
|
1.13 |
ENST00000373759.1
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr19_+_507487
Show fit
|
1.12 |
ENST00000359315.6
|
TPGS1
|
tubulin polyglutamylase complex subunit 1
|
|
chr1_+_25272527
Show fit
|
1.12 |
ENST00000342055.9
ENST00000357542.8
ENST00000417538.6
ENST00000423810.6
ENST00000568195.5
|
RHD
|
Rh blood group D antigen
|
|
chr2_-_288759
Show fit
|
1.11 |
ENST00000452023.1
|
ALKAL2
|
ALK and LTK ligand 2
|
|
chr7_-_38354517
Show fit
|
1.10 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4
|
|
chr19_-_54364908
Show fit
|
1.09 |
ENST00000391742.7
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1
|
|
chr16_-_31094890
Show fit
|
1.09 |
ENST00000532364.1
ENST00000529564.1
ENST00000319788.11
ENST00000354895.4
|
ENSG00000255439.6
VKORC1
|
novel protein, VKORC1 and PRSS53 readthrough
vitamin K epoxide reductase complex subunit 1
|
|
chr1_+_224356852
Show fit
|
1.08 |
ENST00000366858.7
ENST00000366857.9
ENST00000465271.6
ENST00000366856.3
|
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4
|
|
chr16_+_57372481
Show fit
|
1.06 |
ENST00000006053.7
|
CX3CL1
|
C-X3-C motif chemokine ligand 1
|
|
chr20_+_32358303
Show fit
|
1.02 |
ENST00000651418.1
ENST00000375687.10
ENST00000542461.5
ENST00000613218.4
ENST00000646367.1
ENST00000620121.4
|
ASXL1
|
ASXL transcriptional regulator 1
|
|
chr1_-_12618198
Show fit
|
1.01 |
ENST00000616661.5
|
DHRS3
|
dehydrogenase/reductase 3
|
|
chr16_-_31094727
Show fit
|
1.00 |
ENST00000300851.10
ENST00000394975.3
|
VKORC1
|
vitamin K epoxide reductase complex subunit 1
|
|
chr1_-_45340080
Show fit
|
1.00 |
ENST00000354383.10
ENST00000355498.6
ENST00000531105.5
|
MUTYH
|
mutY DNA glycosylase
|
|
chr1_+_32180044
Show fit
|
0.99 |
ENST00000373609.1
|
TXLNA
|
taxilin alpha
|
|
chr4_-_10116324
Show fit
|
0.99 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1
|
|
chr16_+_57372465
Show fit
|
0.99 |
ENST00000563383.1
|
CX3CL1
|
C-X3-C motif chemokine ligand 1
|
|
chr19_+_19385919
Show fit
|
0.99 |
ENST00000417582.6
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr9_-_113275560
Show fit
|
0.98 |
ENST00000374206.4
|
CDC26
|
cell division cycle 26
|
|
chr1_+_113905290
Show fit
|
0.97 |
ENST00000650450.2
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr15_+_88621290
Show fit
|
0.96 |
ENST00000332810.4
ENST00000559528.1
|
AEN
|
apoptosis enhancing nuclease
|
|
chr10_+_46375645
Show fit
|
0.96 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1
|
|
chr22_+_37051731
Show fit
|
0.95 |
ENST00000610767.4
ENST00000402077.7
ENST00000403888.7
|
KCTD17
|
potassium channel tetramerization domain containing 17
|
|
chr1_+_113905156
Show fit
|
0.95 |
ENST00000650596.1
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr17_-_80220325
Show fit
|
0.93 |
ENST00000326317.11
ENST00000570427.1
ENST00000570923.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr11_-_44950731
Show fit
|
0.91 |
ENST00000528473.5
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr14_-_74612021
Show fit
|
0.90 |
ENST00000556690.5
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr19_-_40425982
Show fit
|
0.88 |
ENST00000357949.5
|
SERTAD1
|
SERTA domain containing 1
|
|
chr2_+_197500398
Show fit
|
0.88 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough
|
|
chr1_-_45340381
Show fit
|
0.86 |
ENST00000412971.5
ENST00000372110.7
ENST00000372098.7
ENST00000672818.3
ENST00000529984.5
ENST00000372115.7
|
MUTYH
|
mutY DNA glycosylase
|
|
chr1_-_93180261
Show fit
|
0.86 |
ENST00000370280.1
ENST00000479918.5
|
TMED5
|
transmembrane p24 trafficking protein 5
|
|
chr14_-_36521149
Show fit
|
0.86 |
ENST00000518149.5
|
NKX2-1
|
NK2 homeobox 1
|
|
chr2_-_288056
Show fit
|
0.86 |
ENST00000403610.9
|
ALKAL2
|
ALK and LTK ligand 2
|
|
chr17_-_41525326
Show fit
|
0.86 |
ENST00000593096.1
|
KRT19
|
keratin 19
|
|
chr20_-_23637933
Show fit
|
0.85 |
ENST00000398411.5
|
CST3
|
cystatin C
|
|
chr2_+_32357009
Show fit
|
0.84 |
ENST00000421745.6
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr15_-_34343112
Show fit
|
0.84 |
ENST00000557912.1
ENST00000328848.6
|
NOP10
|
NOP10 ribonucleoprotein
|
|
chr20_+_63658286
Show fit
|
0.83 |
ENST00000360203.11
ENST00000508582.7
ENST00000318100.9
ENST00000356810.5
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr1_-_113905020
Show fit
|
0.83 |
ENST00000432415.5
ENST00000369571.2
ENST00000256658.8
ENST00000369564.5
|
AP4B1
|
adaptor related protein complex 4 subunit beta 1
|
|
chr7_+_75395629
Show fit
|
0.83 |
ENST00000323819.7
ENST00000430211.5
|
TRIM73
|
tripartite motif containing 73
|
|
chr17_+_79074822
Show fit
|
0.83 |
ENST00000311595.14
ENST00000579016.6
|
ENGASE
|
endo-beta-N-acetylglucosaminidase
|
|
chr1_-_54053394
Show fit
|
0.81 |
ENST00000452421.5
ENST00000420738.5
ENST00000440019.5
|
TMEM59
|
transmembrane protein 59
|
|
chr19_-_51027662
Show fit
|
0.80 |
ENST00000594768.5
|
KLK11
|
kallikrein related peptidase 11
|
|
chr4_-_128288163
Show fit
|
0.79 |
ENST00000512483.5
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr1_-_45339995
Show fit
|
0.78 |
ENST00000488731.6
ENST00000435155.1
|
MUTYH
|
mutY DNA glycosylase
|
|
chr22_-_17774412
Show fit
|
0.78 |
ENST00000342111.9
ENST00000622694.5
|
BID
|
BH3 interacting domain death agonist
|
|
chr20_-_23637947
Show fit
|
0.77 |
ENST00000376925.8
|
CST3
|
cystatin C
|
|
chr3_-_58210961
Show fit
|
0.77 |
ENST00000486455.5
ENST00000394549.7
|
DNASE1L3
|
deoxyribonuclease 1 like 3
|
|
chr14_+_24232921
Show fit
|
0.76 |
ENST00000557854.5
ENST00000399440.7
ENST00000559104.5
ENST00000456667.7
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr7_+_116526277
Show fit
|
0.76 |
ENST00000393468.1
ENST00000393467.1
|
CAV1
|
caveolin 1
|
|
chrX_+_40580894
Show fit
|
0.75 |
ENST00000636409.1
ENST00000637327.1
ENST00000637526.1
ENST00000638153.1
ENST00000378438.9
ENST00000636970.1
ENST00000636196.1
ENST00000636251.1
ENST00000637482.1
ENST00000636580.2
ENST00000423649.2
ENST00000636287.1
|
ATP6AP2
|
ATPase H+ transporting accessory protein 2
|
|
chr2_-_74529670
Show fit
|
0.75 |
ENST00000377526.4
|
AUP1
|
AUP1 lipid droplet regulating VLDL assembly factor
|
|
chr16_-_31094549
Show fit
|
0.75 |
ENST00000394971.7
|
VKORC1
|
vitamin K epoxide reductase complex subunit 1
|
|
chr4_+_26857678
Show fit
|
0.73 |
ENST00000494628.6
|
STIM2
|
stromal interaction molecule 2
|
|
chrX_+_40581035
Show fit
|
0.73 |
ENST00000447485.6
|
ATP6AP2
|
ATPase H+ transporting accessory protein 2
|
|
chr19_-_43754901
Show fit
|
0.72 |
ENST00000270066.11
ENST00000601170.5
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor
|
|
chr12_-_113335030
Show fit
|
0.72 |
ENST00000552014.5
ENST00000680972.1
ENST00000548186.5
ENST00000202831.7
ENST00000549181.5
|
SLC8B1
|
solute carrier family 8 member B1
|
|
chr12_+_121132869
Show fit
|
0.72 |
ENST00000328963.10
|
P2RX7
|
purinergic receptor P2X 7
|
|
chr14_-_74612226
Show fit
|
0.72 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr16_+_88382950
Show fit
|
0.70 |
ENST00000565624.3
|
ZNF469
|
zinc finger protein 469
|
|
chrX_+_71254781
Show fit
|
0.70 |
ENST00000677446.1
|
NONO
|
non-POU domain containing octamer binding
|
|
chr1_-_153608136
Show fit
|
0.70 |
ENST00000368703.6
|
S100A16
|
S100 calcium binding protein A16
|
|
chr20_+_36605820
Show fit
|
0.68 |
ENST00000342422.3
|
RAB5IF
|
RAB5 interacting factor
|
|
chr22_-_17774482
Show fit
|
0.67 |
ENST00000399765.5
ENST00000614949.4
ENST00000399767.6
|
BID
|
BH3 interacting domain death agonist
|
|
chr22_+_37051787
Show fit
|
0.67 |
ENST00000456470.1
|
KCTD17
|
potassium channel tetramerization domain containing 17
|
|
chr1_-_54053192
Show fit
|
0.66 |
ENST00000371337.3
ENST00000234831.10
|
TMEM59
|
transmembrane protein 59
|
|
chr14_-_36320582
Show fit
|
0.65 |
ENST00000604336.5
ENST00000359527.11
ENST00000621657.4
ENST00000603139.1
ENST00000318473.11
ENST00000416007.9
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr12_+_6828377
Show fit
|
0.65 |
ENST00000290510.10
|
P3H3
|
prolyl 3-hydroxylase 3
|
|
chr2_+_109129199
Show fit
|
0.64 |
ENST00000309415.8
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr11_+_64359142
Show fit
|
0.63 |
ENST00000528057.5
ENST00000334205.9
|
RPS6KA4
|
ribosomal protein S6 kinase A4
|
|
chr15_-_22980334
Show fit
|
0.63 |
ENST00000610365.4
ENST00000617928.5
ENST00000611832.4
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr11_+_45805108
Show fit
|
0.62 |
ENST00000530471.1
ENST00000314134.4
|
SLC35C1
|
solute carrier family 35 member C1
|
|
chr13_+_51584435
Show fit
|
0.59 |
ENST00000612477.1
ENST00000298125.7
|
WDFY2
|
WD repeat and FYVE domain containing 2
|
|
chr17_+_78214186
Show fit
|
0.59 |
ENST00000301633.8
ENST00000350051.8
ENST00000374948.6
ENST00000590449.1
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr5_+_122312164
Show fit
|
0.59 |
ENST00000514497.6
ENST00000261367.11
|
SNCAIP
|
synuclein alpha interacting protein
|
|
chr14_-_50532518
Show fit
|
0.58 |
ENST00000682126.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
|
chr4_-_48906720
Show fit
|
0.58 |
ENST00000381464.6
ENST00000508632.6
|
OCIAD2
|
OCIA domain containing 2
|
|
chr14_-_64504570
Show fit
|
0.58 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr1_-_54406385
Show fit
|
0.57 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr5_+_122311740
Show fit
|
0.57 |
ENST00000506272.5
ENST00000508681.5
ENST00000509154.6
|
SNCAIP
|
synuclein alpha interacting protein
|
|
chr11_+_64359199
Show fit
|
0.56 |
ENST00000530504.1
|
RPS6KA4
|
ribosomal protein S6 kinase A4
|
|
chr1_-_93180377
Show fit
|
0.56 |
ENST00000370282.8
|
TMED5
|
transmembrane p24 trafficking protein 5
|
|
chr8_+_119416427
Show fit
|
0.55 |
ENST00000259526.4
|
CCN3
|
cellular communication network factor 3
|
|
chr17_+_28371656
Show fit
|
0.55 |
ENST00000585482.6
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr14_-_50532590
Show fit
|
0.55 |
ENST00000013125.9
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
|
chr22_+_40370578
Show fit
|
0.55 |
ENST00000457767.5
ENST00000248929.14
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr4_+_2536630
Show fit
|
0.54 |
ENST00000637812.2
|
FAM193A
|
family with sequence similarity 193 member A
|
|
chr1_-_53945661
Show fit
|
0.54 |
ENST00000194214.10
|
HSPB11
|
heat shock protein family B (small) member 11
|
|
chr22_+_37906275
Show fit
|
0.54 |
ENST00000215957.10
ENST00000445494.6
ENST00000680578.1
ENST00000424008.2
|
MICALL1
|
MICAL like 1
|
|
chr20_+_36605734
Show fit
|
0.53 |
ENST00000344795.8
ENST00000373852.9
|
RAB5IF
|
RAB5 interacting factor
|
|
chr13_+_38349822
Show fit
|
0.53 |
ENST00000379649.5
ENST00000239878.9
|
UFM1
|
ubiquitin fold modifier 1
|
|
chr1_-_54053461
Show fit
|
0.52 |
ENST00000371341.5
|
TMEM59
|
transmembrane protein 59
|
|
chr16_+_51553436
Show fit
|
0.52 |
ENST00000565308.2
|
HNRNPA1P48
|
heterogeneous nuclear ribonucleoprotein A1 pseudogene 48
|
|
chr11_-_65540610
Show fit
|
0.51 |
ENST00000532661.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|