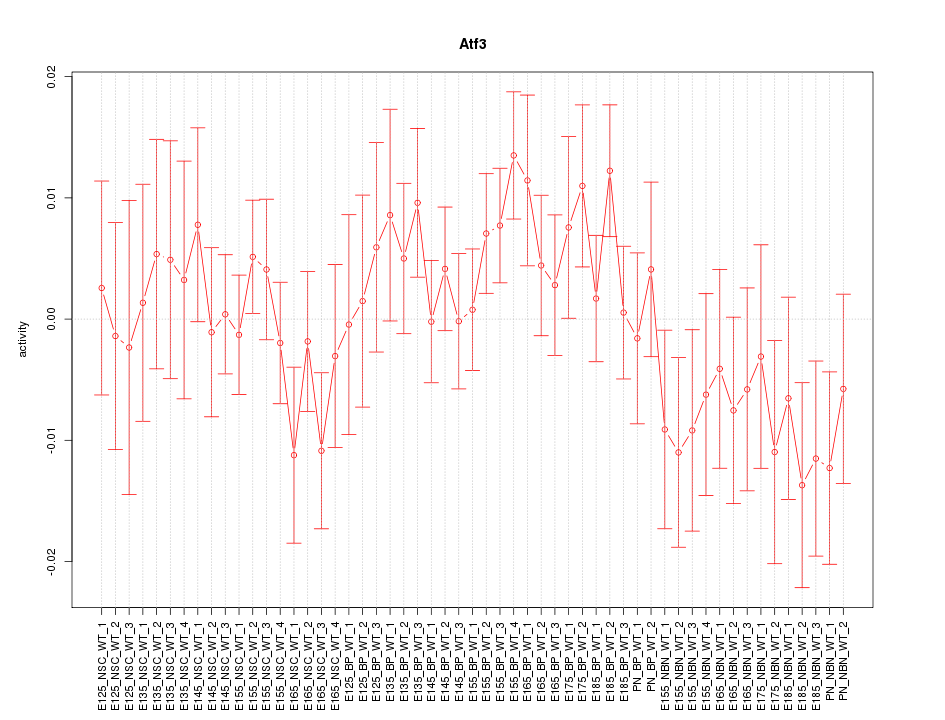
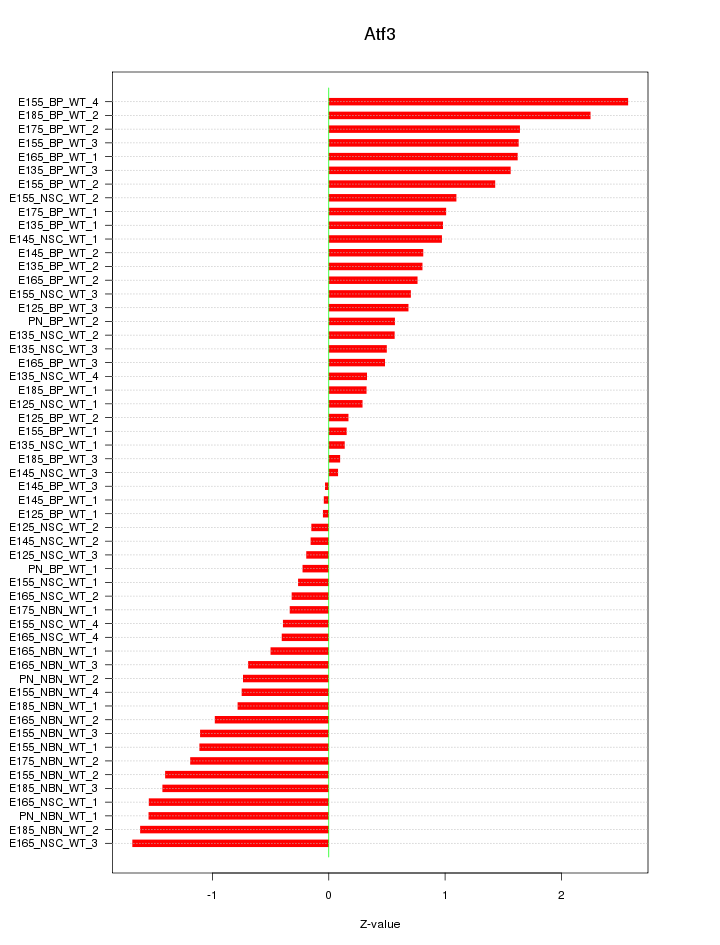
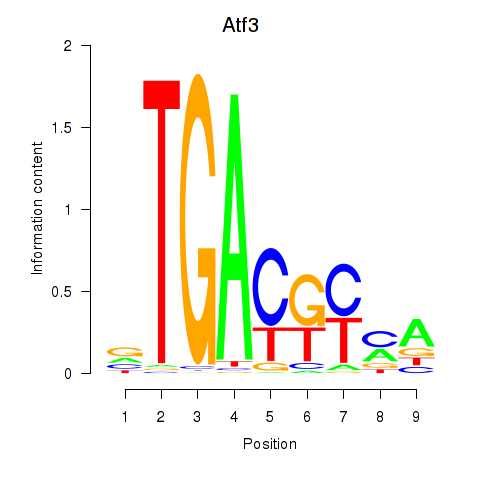
Motif ID: Atf3
Z-value: 1.000
Transcription factors associated with Atf3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atf3 | ENSMUSG00000026628.8 | Atf3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf3 | mm10_v2_chr1_-_191183244_191183340 | -0.41 | 2.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 2.5 | 7.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 1.5 | 6.1 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 1.3 | 3.8 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.2 | 3.5 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 1.1 | 4.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 1.1 | 5.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 2.7 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.8 | 2.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.7 | 4.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.7 | 2.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.7 | 2.6 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.6 | 3.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 | 5.8 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.6 | 4.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.6 | 1.7 | GO:1901079 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) |
| 0.6 | 2.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.6 | 3.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.6 | 1.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 2.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.5 | 3.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 1.6 | GO:0046370 | fructose biosynthetic process(GO:0046370) |
| 0.5 | 1.5 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.5 | 1.4 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.5 | 2.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.5 | 2.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.4 | 1.3 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.4 | 2.5 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.4 | 1.6 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.4 | 0.8 | GO:1900145 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.4 | 2.4 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 2.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.4 | 1.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 | 1.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.4 | 1.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.4 | 3.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 3.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.4 | 1.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 1.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.4 | 1.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.3 | 0.7 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.3 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.3 | 1.7 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.3 | 2.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 | 1.0 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 0.7 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.3 | 1.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 1.0 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.3 | 0.7 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.3 | 1.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 1.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.3 | 9.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 1.3 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.3 | 2.9 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 2.9 | GO:0030432 | peristalsis(GO:0030432) |
| 0.3 | 1.2 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.3 | 0.9 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.3 | 3.0 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.3 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.3 | 1.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.3 | 1.5 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.3 | 1.2 | GO:0000237 | leptotene(GO:0000237) |
| 0.3 | 0.9 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 3.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.3 | 1.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 2.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 0.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.3 | 1.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.3 | 1.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 2.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 | 1.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 2.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.3 | 1.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 0.8 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.3 | 9.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 2.4 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.3 | 1.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 0.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.3 | 0.8 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 2.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 0.8 | GO:0090403 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) oxidative stress-induced premature senescence(GO:0090403) |
| 0.3 | 1.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.3 | 0.8 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 0.7 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.2 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 2.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.7 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 0.7 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.2 | 1.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 3.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.7 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 0.7 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.9 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.2 | 1.1 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.2 | 0.4 | GO:1902566 | regulation of eosinophil activation(GO:1902566) |
| 0.2 | 0.9 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 1.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 1.3 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 1.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 3.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 0.2 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.2 | 1.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.8 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.2 | 1.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 1.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 1.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 1.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 1.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 0.7 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 0.6 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 1.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 0.5 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.2 | 0.5 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 1.3 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.2 | 1.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 2.1 | GO:0072431 | signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.2 | 0.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 1.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 1.5 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 | 1.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 0.5 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.2 | 0.2 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 3.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 0.7 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 2.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 2.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 4.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.2 | 0.5 | GO:0010985 | negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.2 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.7 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.2 | 1.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 0.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.2 | 2.6 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.2 | 0.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.6 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 2.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 0.5 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 | 3.8 | GO:0007140 | male meiosis(GO:0007140) |
| 0.2 | 0.6 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.2 | 0.5 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 1.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 2.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.2 | 0.6 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 0.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.9 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.4 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 0.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.6 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 1.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.1 | 1.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.4 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.1 | 0.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.5 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 1.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.9 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 0.6 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 1.2 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.5 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.1 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.1 | 2.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 1.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.5 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.2 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 0.5 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 0.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.5 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.8 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 2.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.0 | GO:1904714 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.5 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 1.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 2.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.5 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.1 | 0.5 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.7 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.3 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.6 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) |
| 0.1 | 1.2 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.1 | 1.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.1 | 0.2 | GO:0032275 | luteinizing hormone secretion(GO:0032275) |
| 0.1 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.6 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.6 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.1 | 0.5 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 | 0.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.6 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.4 | GO:0090282 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 0.2 | GO:0001996 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.1 | 0.9 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.3 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.8 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.5 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.4 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.1 | 0.3 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.1 | 1.0 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.7 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.3 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.7 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.4 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.2 | GO:1900223 | positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.1 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.3 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 0.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 1.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 1.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.6 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.4 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 2.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.7 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 2.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 2.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 4.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.2 | GO:1902534 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) modulation by virus of host autophagy(GO:0039519) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 1.0 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 3.7 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.1 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.1 | GO:0061055 | myotome development(GO:0061055) regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.1 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.1 | 0.8 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 1.0 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 | 1.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.3 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 0.1 | 0.3 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0045414 | regulation of interleukin-8 biosynthetic process(GO:0045414) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.1 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.2 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.7 | GO:2000192 | negative regulation of fatty acid transport(GO:2000192) |
| 0.1 | 1.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 2.1 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.4 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 0.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 0.1 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.3 | GO:0051189 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.1 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.3 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 1.2 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) Golgi reassembly(GO:0090168) |
| 0.0 | 0.3 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.4 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.6 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 1.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.9 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 2.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.3 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 1.0 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil chemotaxis(GO:0090022) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.5 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) positive regulation of maintenance of sister chromatid cohesion(GO:0034093) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.3 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.3 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.5 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 2.1 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.7 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.3 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.3 | GO:1903363 | negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:1903912 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) regulation of translation initiation in response to endoplasmic reticulum stress(GO:0036491) eiF2alpha phosphorylation in response to endoplasmic reticulum stress(GO:0036492) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:1903750 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0034109 | homotypic cell-cell adhesion(GO:0034109) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 1.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.6 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 1.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.2 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0019048 | modulation by virus of host morphology or physiology(GO:0019048) |
| 0.0 | 0.1 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0006475 | internal protein amino acid acetylation(GO:0006475) |
| 0.0 | 0.2 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:2000489 | hepatic stellate cell activation(GO:0035733) regulation of hepatic stellate cell activation(GO:2000489) |
| 0.0 | 0.0 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.4 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.1 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.0 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0001710 | mesodermal cell fate commitment(GO:0001710) |
| 0.0 | 0.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0042100 | B cell proliferation(GO:0042100) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.8 | 2.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 2.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 2.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.6 | 11.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.6 | 1.7 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.6 | 1.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 3.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.5 | 3.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.5 | 4.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 5.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.5 | 1.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.5 | 1.8 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 1.6 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.4 | 0.8 | GO:0097413 | Lewy body(GO:0097413) Lewy body core(GO:1990037) |
| 0.4 | 3.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 2.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 2.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 1.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 1.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 1.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 1.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 1.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 1.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 0.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 2.0 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 5.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 1.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 4.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 1.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 2.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 2.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 1.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 2.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.8 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.2 | 1.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 1.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 9.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 0.5 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 0.5 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 2.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 8.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.4 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.1 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 2.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.6 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.1 | 1.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 1.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 2.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 10.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.6 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.4 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 1.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 1.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.4 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 9.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.1 | 5.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.9 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 3.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 2.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.7 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.4 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 2.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.7 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 1.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.1 | 3.2 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 1.0 | 2.9 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.9 | 4.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 2.9 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.7 | 2.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.7 | 6.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.6 | 1.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.6 | 1.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.6 | 8.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.5 | 3.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.5 | 2.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.5 | 2.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.5 | 1.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.5 | 2.0 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.5 | 1.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.5 | 2.0 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.4 | 1.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.4 | 5.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 2.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.4 | 1.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 6.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 1.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 1.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.3 | 1.0 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.3 | 1.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.6 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.3 | 1.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.3 | 0.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 0.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 0.9 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.3 | 0.9 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.3 | 5.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 1.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 1.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 0.8 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 1.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 3.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 0.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 2.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 2.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 1.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 2.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 0.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 3.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 2.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 5.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 5.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.2 | 0.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 0.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.2 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.5 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 0.8 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 0.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.6 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 1.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.2 | 1.1 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 2.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 2.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 3.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.1 | 0.4 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 2.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 5.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.4 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 2.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.5 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.6 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 2.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 3.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 1.8 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.1 | 0.3 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 3.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 1.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.3 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.9 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.8 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.2 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 1.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 4.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 2.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.5 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 13.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.9 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.2 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 1.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 2.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.2 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.2 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.1 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.2 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.2 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 4.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 2.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 6.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0055102 | lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0019863 | IgG receptor activity(GO:0019770) IgE binding(GO:0019863) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 1.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.9 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 6.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.8 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 2.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.0 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.8 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.0 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 4.0 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 5.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 6.6 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.3 | 13.6 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.3 | 0.8 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.3 | 0.6 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.2 | 10.7 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.2 | 7.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 7.1 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.7 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.0 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.6 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 1.9 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 3.4 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.1 | 1.2 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.7 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.5 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.8 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.8 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.8 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 2.5 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.1 | 1.4 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.1 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 5.1 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 1.5 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 0.6 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.3 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.3 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.6 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.1 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.6 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.0 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 1.3 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.2 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 1.0 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.2 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.1 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.0 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 12.9 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.4 | 3.6 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 5.8 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.3 | 18.8 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 3.2 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 7.7 | REACTOME_ACTIVATION_OF_ATR_IN_RESPONSE_TO_REPLICATION_STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.3 | 1.1 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.0 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 0.4 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 1.6 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 1.4 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 4.2 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 4.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.2 | 0.9 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 2.2 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.0 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.2 | REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.1 | 4.1 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 3.4 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 4.1 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.6 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.6 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.0 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.5 | REACTOME_DESTABILIZATION_OF_MRNA_BY_BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.0 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.4 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.6 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.8 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.5 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.2 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.6 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.5 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.0 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.4 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.3 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.4 | REACTOME_TAK1_ACTIVATES_NFKB_BY_PHOSPHORYLATION_AND_ACTIVATION_OF_IKKS_COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 1.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.7 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.6 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.3 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 0.8 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 4.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.3 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.2 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.5 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 2.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.9 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.2 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.7 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.3 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.2 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.2 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.9 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME_TRNA_AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.8 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME_CHROMOSOME_MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 0.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.2 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_CYCLIN_B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.4 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |