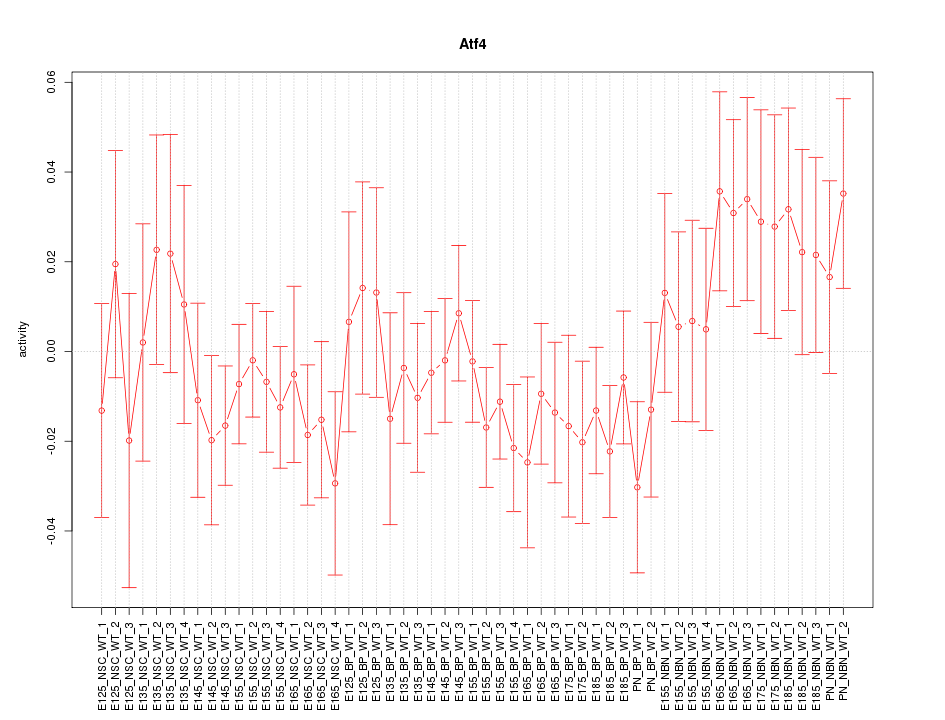
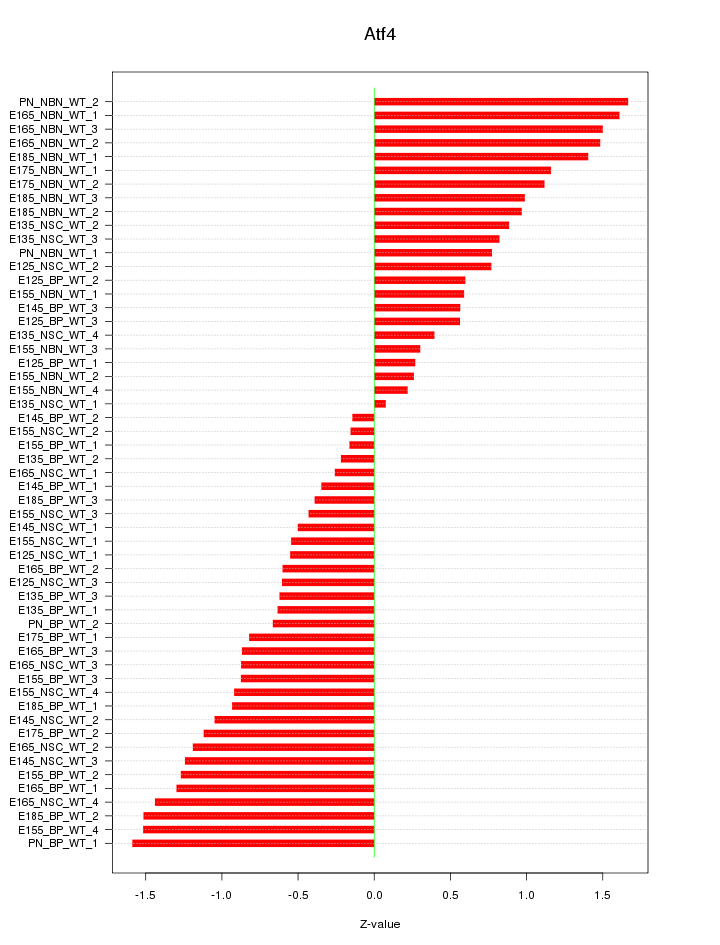
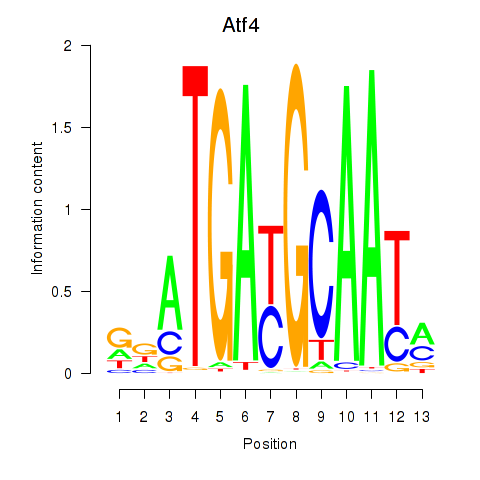
Motif ID: Atf4
Z-value: 0.921
Transcription factors associated with Atf4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atf4 | ENSMUSG00000042406.7 | Atf4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf4 | mm10_v2_chr15_+_80255184_80255263 | -0.16 | 2.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) transforming growth factor-beta secretion(GO:0038044) |
| 1.4 | 5.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.4 | 6.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.1 | 4.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.8 | 2.5 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.8 | 2.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.7 | 2.9 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.6 | 5.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.6 | 2.8 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.5 | 1.6 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.4 | 1.6 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 1.6 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.4 | 1.5 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.4 | 1.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 2.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 2.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 1.6 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 1.5 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.3 | 1.2 | GO:1904706 | heme oxidation(GO:0006788) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 | 0.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.3 | 0.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 1.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 3.8 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.2 | 0.5 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.2 | 1.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 1.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 4.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 1.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 1.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.2 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.8 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 1.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.5 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.7 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.5 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 1.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 1.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 0.2 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 | 0.9 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 3.8 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 1.5 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 5.9 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 2.9 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 2.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 1.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.3 | 5.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 1.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 5.5 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 6.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 5.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 6.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 3.5 | GO:0098793 | presynapse(GO:0098793) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 1.1 | 5.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.8 | 2.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.6 | 2.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 1.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 4.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.5 | 1.6 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.5 | 3.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 1.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.4 | 1.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.4 | 1.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 2.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 1.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 0.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 1.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 0.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 1.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.8 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 6.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 2.8 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.2 | 1.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.5 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 5.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 5.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 6.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) proteasome binding(GO:0070628) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.2 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.0 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.9 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.1 | 0.8 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.4 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.2 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.9 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 2.7 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.9 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 11.0 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 2.9 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 8.1 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 5.9 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME_RAF_MAP_KINASE_CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.4 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.6 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.2 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.8 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.9 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.1 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |