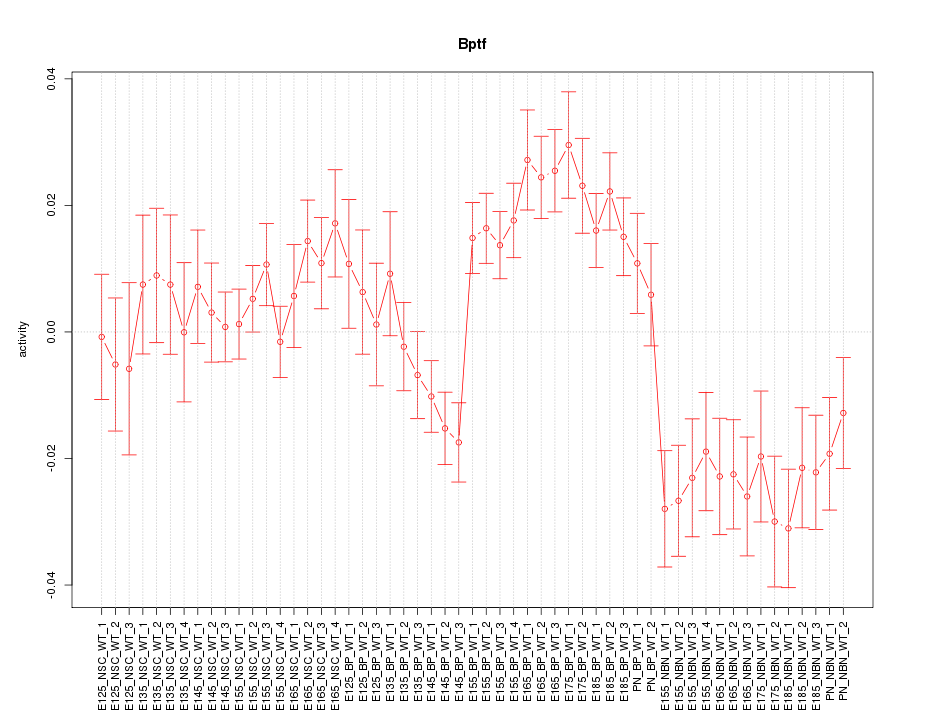
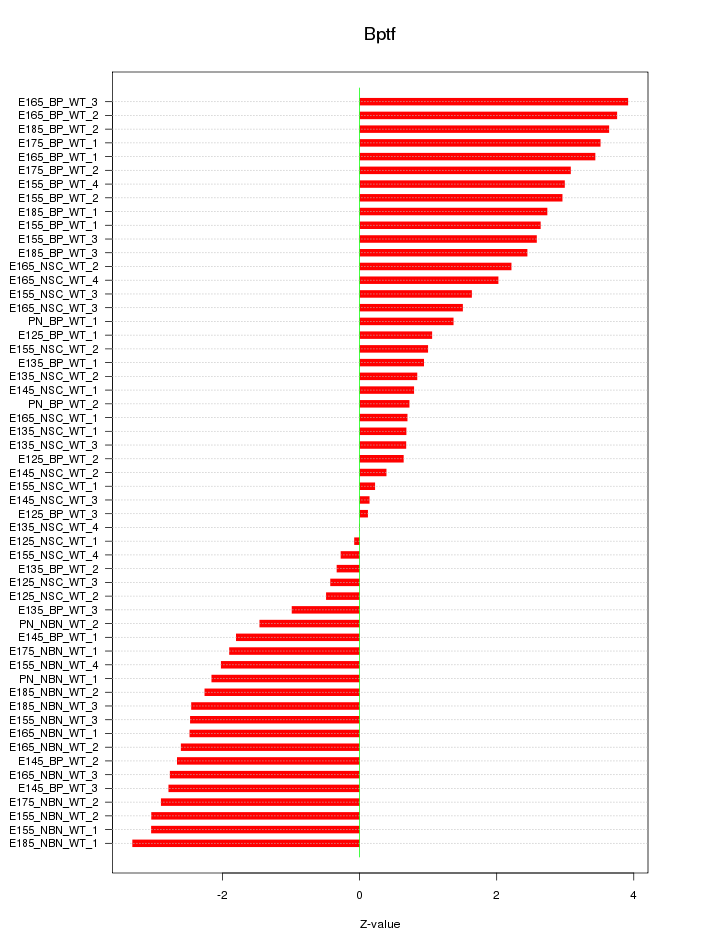
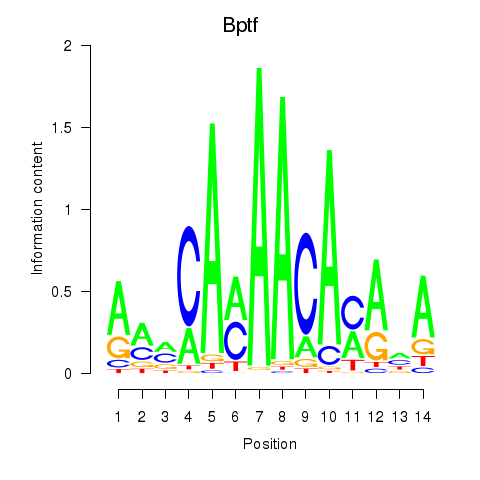
Motif ID: Bptf
Z-value: 2.149
Transcription factors associated with Bptf:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Bptf | ENSMUSG00000040481.10 | Bptf |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bptf | mm10_v2_chr11_-_107131922_107131954 | 0.51 | 6.1e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 6.4 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 2.9 | 14.4 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 2.6 | 7.9 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 2.6 | 7.9 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 2.6 | 7.8 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 2.4 | 4.9 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) |
| 2.2 | 6.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 2.2 | 10.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.1 | 33.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 2.1 | 2.1 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 1.9 | 5.8 | GO:0046544 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 1.9 | 5.8 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) positive regulation of NK T cell activation(GO:0051135) |
| 1.9 | 5.7 | GO:0030421 | defecation(GO:0030421) |
| 1.8 | 7.4 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 1.8 | 3.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.7 | 13.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.7 | 5.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.6 | 4.8 | GO:0060023 | soft palate development(GO:0060023) |
| 1.4 | 7.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 1.4 | 4.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.4 | 11.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 1.2 | 2.4 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 1.2 | 10.5 | GO:0048102 | autophagic cell death(GO:0048102) |
| 1.1 | 2.2 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 1.1 | 3.2 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 1.1 | 3.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 1.0 | 1.0 | GO:0072284 | metanephric S-shaped body morphogenesis(GO:0072284) |
| 1.0 | 4.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.0 | 3.9 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.9 | 2.8 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 2.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.9 | 13.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.9 | 8.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.9 | 9.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.8 | 5.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.8 | 7.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.8 | 2.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.8 | 32.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.8 | 2.4 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.8 | 3.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.8 | 2.3 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.7 | 17.9 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.7 | 9.0 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.7 | 2.2 | GO:1905065 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.7 | 2.2 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.7 | 7.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.7 | 3.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.7 | 0.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.7 | 4.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.7 | 6.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 8.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.7 | 4.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.7 | 9.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.7 | 4.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.7 | 2.0 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.7 | 2.6 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.6 | 5.7 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.6 | 1.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.6 | 1.8 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.6 | 1.8 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.6 | 3.5 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.6 | 1.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.6 | 5.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.6 | 2.9 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.6 | 1.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.5 | 1.6 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.5 | 3.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.5 | 3.2 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.5 | 4.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.5 | 2.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.5 | 2.1 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.5 | 2.1 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.5 | 5.2 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.5 | 2.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.5 | 7.6 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.5 | 2.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.5 | 3.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 1.5 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.5 | 1.5 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.5 | 2.9 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.5 | 3.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.5 | 4.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 | 1.4 | GO:0051095 | regulation of helicase activity(GO:0051095) |
| 0.5 | 1.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.5 | 1.9 | GO:1990414 | negative regulation of mitotic recombination(GO:0045950) replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.5 | 6.0 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.5 | 1.4 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.5 | 1.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 1.8 | GO:0060032 | notochord regression(GO:0060032) |
| 0.4 | 1.3 | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) |
| 0.4 | 0.9 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.4 | 2.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.4 | 1.3 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.4 | 1.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.4 | 2.2 | GO:2000482 | interleukin-8 secretion(GO:0072606) regulation of interleukin-8 secretion(GO:2000482) |
| 0.4 | 1.3 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.4 | 2.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 4.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.4 | 1.6 | GO:0014012 | negative regulation of Schwann cell proliferation(GO:0010626) peripheral nervous system axon regeneration(GO:0014012) |
| 0.4 | 3.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 1.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 3.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 1.5 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.4 | 3.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 | 1.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.4 | 2.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.4 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 1.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 6.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.4 | 2.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 0.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.4 | 12.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.4 | 1.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.4 | 10.6 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 1.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 0.3 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.3 | 9.2 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.3 | 4.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.3 | 10.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.3 | 2.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 1.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 2.7 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.3 | 1.7 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 | 1.3 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.3 | 1.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.3 | 1.0 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.3 | 1.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.3 | 3.2 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.3 | 2.2 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.3 | 2.5 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.3 | 9.0 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.3 | 1.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.3 | 1.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 0.3 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.3 | 4.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 0.9 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.3 | 2.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 1.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 4.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 1.4 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.3 | 0.8 | GO:0015793 | glycerol transport(GO:0015793) urea transport(GO:0015840) |
| 0.3 | 0.6 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.3 | 2.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.3 | 0.8 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 1.9 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.3 | 2.2 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.3 | 1.1 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.3 | 1.9 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 2.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.3 | 1.1 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.3 | 5.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 2.8 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.3 | 0.8 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.3 | 1.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.3 | 0.8 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.3 | 1.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.3 | 1.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 1.5 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 2.0 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 1.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 4.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.7 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.2 | 0.9 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.2 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 0.7 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 1.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 4.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.9 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 1.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 3.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.9 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 2.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.2 | 0.7 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.2 | 0.7 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) |
| 0.2 | 0.9 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 0.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 1.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.2 | 1.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.4 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.2 | 0.9 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 0.6 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 11.7 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.2 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 5.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 4.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 0.6 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 2.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.6 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 0.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 3.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 1.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.6 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.6 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.2 | 1.5 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.2 | 3.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 7.8 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.2 | 3.3 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.2 | 1.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 1.9 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 0.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.7 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 0.5 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 0.9 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 0.7 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.2 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 2.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.2 | 0.7 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 2.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 1.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 1.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.6 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 2.6 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 0.3 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) |
| 0.2 | 5.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 1.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 3.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.2 | 1.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 2.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.2 | 1.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 1.7 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) fear response(GO:0042596) |
| 0.2 | 0.5 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.1 | 6.5 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 1.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.2 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) |
| 0.1 | 1.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 0.4 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.1 | 0.8 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.8 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.4 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 2.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 1.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 2.8 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 2.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 4.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.4 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.2 | GO:0035729 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 1.3 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 1.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 2.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.2 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.1 | 1.1 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.6 | GO:0098706 | ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 2.9 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 2.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.8 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) |
| 0.1 | 0.8 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 1.0 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 2.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.5 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 1.7 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 0.9 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.4 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 1.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 2.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.3 | GO:0042939 | glutathione transport(GO:0034635) xenobiotic transport(GO:0042908) tripeptide transport(GO:0042939) |
| 0.1 | 1.7 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.3 | GO:1905077 | T-helper 17 cell lineage commitment(GO:0072540) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 0.5 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.8 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 4.1 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 4.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.5 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.1 | 3.6 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.8 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 1.1 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 1.4 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.1 | 4.3 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.9 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.4 | GO:1903363 | negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.1 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.6 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 0.6 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 3.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 1.0 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.8 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.1 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.8 | GO:0000578 | embryonic axis specification(GO:0000578) |
| 0.1 | 3.4 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 2.0 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.1 | 1.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.0 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 1.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.0 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 5.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:0045743 | positive regulation of vascular permeability(GO:0043117) positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.9 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 8.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.6 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.1 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 | 1.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 1.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.4 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.1 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 5.3 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0070243 | regulation of thymocyte apoptotic process(GO:0070243) |
| 0.0 | 0.3 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.7 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.3 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 4.4 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 1.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 2.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.7 | GO:0065004 | nucleosome organization(GO:0034728) protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.2 | GO:0015819 | lysine transport(GO:0015819) ornithine transport(GO:0015822) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 1.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 3.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 1.2 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.4 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 1.1 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 2.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.2 | GO:0010453 | regulation of cell fate commitment(GO:0010453) |
| 0.0 | 0.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 1.1 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.6 | GO:0043038 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.0 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.9 | 7.8 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.4 | 11.5 | GO:0005818 | aster(GO:0005818) |
| 1.4 | 13.6 | GO:0000796 | condensin complex(GO:0000796) |
| 1.3 | 3.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.2 | 11.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.8 | 6.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.8 | 2.4 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.8 | 15.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.6 | 1.8 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.6 | 1.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.6 | 9.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.6 | 3.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.5 | 4.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.5 | 3.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 5.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 4.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.5 | 9.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.4 | 2.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 7.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.4 | 2.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.4 | 1.2 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.4 | 2.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 3.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.4 | 6.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 4.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 6.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 6.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 4.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 3.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.3 | 1.3 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.3 | 9.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 1.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 0.9 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 1.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 3.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 10.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 2.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 6.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 6.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 12.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 5.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 3.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 8.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 1.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 6.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 10.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 0.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 4.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.7 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.2 | 0.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 3.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.0 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 2.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 6.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.9 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.4 | GO:0030690 | Noc complex(GO:0030689) Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 1.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 7.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 4.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 17.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 5.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.4 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 8.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 13.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 2.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 2.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 2.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.4 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 10.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 4.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 4.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 1.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 3.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 70.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 3.5 | 10.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 3.0 | 9.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 2.9 | 20.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 2.8 | 8.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 2.5 | 7.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.9 | 7.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.9 | 5.8 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 1.6 | 4.9 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.6 | 4.7 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.5 | 4.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 1.5 | 8.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.4 | 4.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.4 | 12.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.3 | 5.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.3 | 7.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.9 | 2.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 4.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.9 | 2.6 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.9 | 4.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.9 | 2.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.9 | 12.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 6.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.8 | 5.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.8 | 6.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.7 | 2.9 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.7 | 9.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.7 | 3.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 2.8 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.7 | 2.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.7 | 4.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.7 | 6.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.7 | 2.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.7 | 3.3 | GO:0047134 | thioredoxin-disulfide reductase activity(GO:0004791) protein-disulfide reductase activity(GO:0047134) |
| 0.6 | 4.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 1.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.6 | 7.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.6 | 4.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.6 | 1.8 | GO:0016774 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) phosphotransferase activity, carboxyl group as acceptor(GO:0016774) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.6 | 4.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.6 | 4.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 9.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 3.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.6 | 10.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.6 | 2.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.5 | 1.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.5 | 6.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.5 | 1.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.5 | 5.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.5 | 4.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.5 | 2.3 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.5 | 8.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 2.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 1.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.4 | 7.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.4 | 1.9 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 3.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.4 | 1.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.4 | 10.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 2.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 1.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 3.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.3 | 1.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 7.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 10.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 1.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 4.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 4.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 2.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 4.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 4.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 0.9 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 2.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 12.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 2.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.3 | 1.1 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 1.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 1.1 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.3 | 1.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 1.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 2.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 3.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 1.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 1.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 6.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 1.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 1.4 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 0.9 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 5.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 2.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.2 | 1.6 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.2 | 0.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 1.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 2.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 0.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 0.8 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 1.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 4.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 0.9 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 2.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 0.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 6.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 30.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 4.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 2.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 1.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 2.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 2.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 6.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 1.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 1.6 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) chemorepellent activity(GO:0045499) |
| 0.2 | 3.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 5.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 2.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.2 | 2.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.0 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 4.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 6.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.3 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016796) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 2.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 5.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 5.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 21.2 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 2.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 7.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 3.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 5.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 8.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 6.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.5 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 3.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 5.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 13.3 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 0.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 5.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 2.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 1.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 2.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 2.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 31.8 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 2.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 6.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 2.5 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.6 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 1.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.2 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 2.7 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.8 | 7.5 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.6 | 26.6 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.6 | 23.4 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.6 | 4.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 23.0 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.4 | 24.0 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 4.1 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.3 | 5.6 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.3 | 4.3 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.2 | 2.4 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 6.1 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 5.7 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 33.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 4.1 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.3 | PID_HIV_NEF_PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 9.5 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.2 | 3.3 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.2 | 10.2 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 1.2 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.2 | 3.5 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 6.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.2 | 2.8 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 12.6 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.2 | 4.7 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.2 | 6.0 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 3.9 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 11.5 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.2 | 7.2 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 2.2 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.2 | 2.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 8.2 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 0.5 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.7 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.2 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.0 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 2.4 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.8 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.8 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.3 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.3 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.5 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 4.7 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.7 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.1 | 1.5 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.5 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.2 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 0.8 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.9 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 3.9 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.5 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.3 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.9 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.0 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 2.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 1.9 | 18.8 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.9 | 3.5 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.7 | 0.7 | REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.6 | 9.9 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.6 | 7.4 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.5 | 5.8 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 4.3 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 5.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 7.0 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.4 | 4.1 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 9.7 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 4.3 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 8.9 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.3 | 1.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 15.6 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 2.9 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 1.3 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 0.8 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 6.9 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 7.9 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 4.1 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 2.4 | REACTOME_DESTABILIZATION_OF_MRNA_BY_BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.2 | 1.3 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 13.9 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 4.2 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 1.7 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 13.8 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.1 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 4.7 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.1 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.5 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.3 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.7 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.3 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.4 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 4.1 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.5 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.7 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.7 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.8 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.2 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.8 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 2.2 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.6 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 3.2 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.7 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.7 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.3 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.0 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.5 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.1 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.9 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.1 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 2.4 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.4 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.3 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 2.7 | REACTOME_INTERFERON_SIGNALING | Genes involved in Interferon Signaling |
| 0.0 | 1.2 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.1 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME_LIGAND_GATED_ION_CHANNEL_TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |