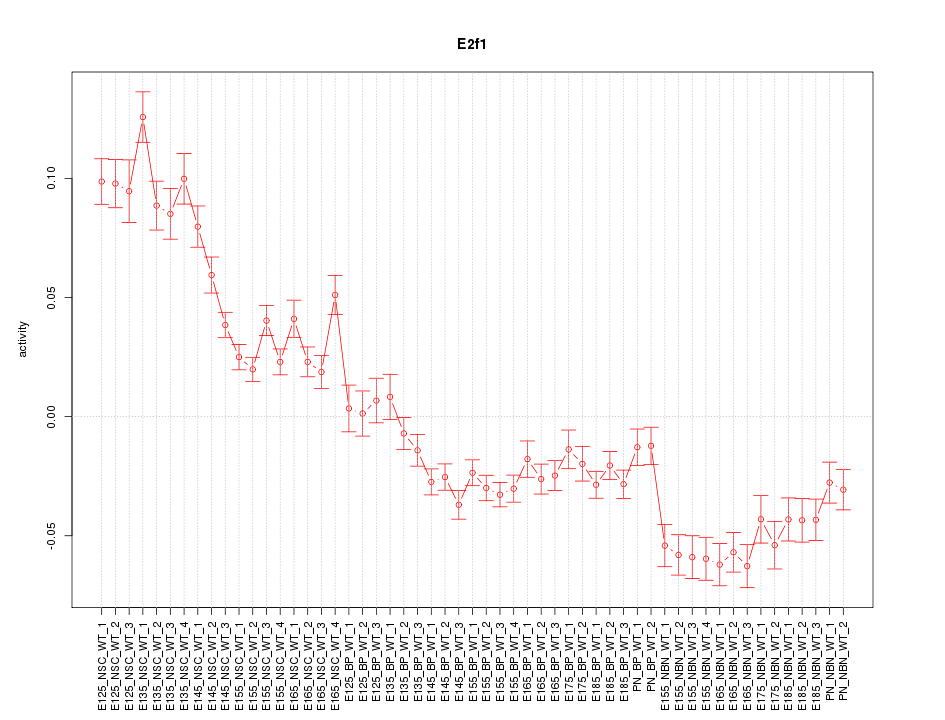
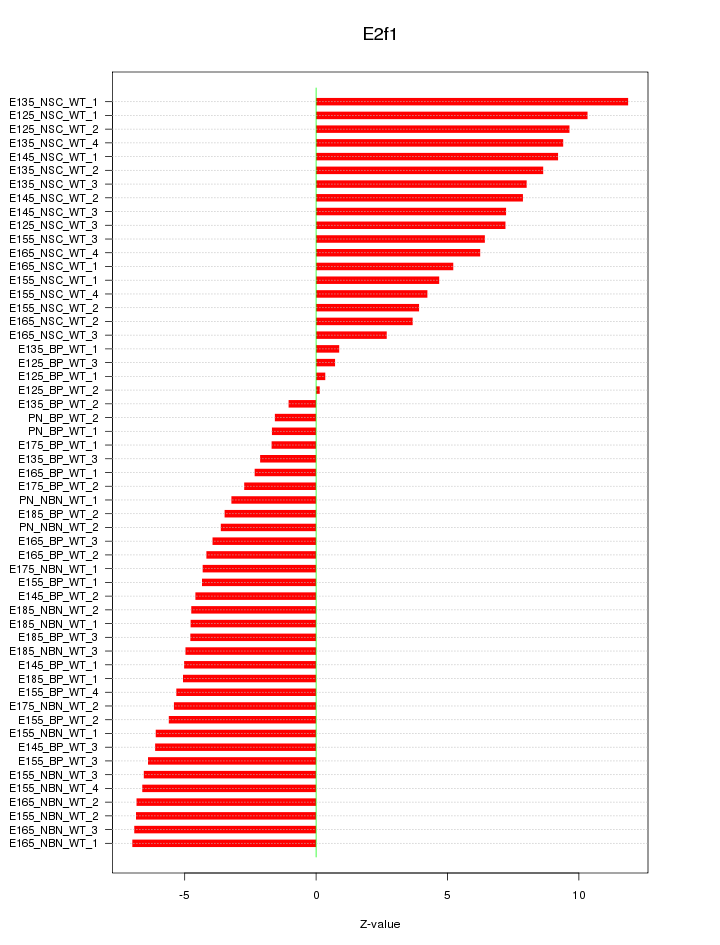
Motif ID: E2f1
Z-value: 5.694
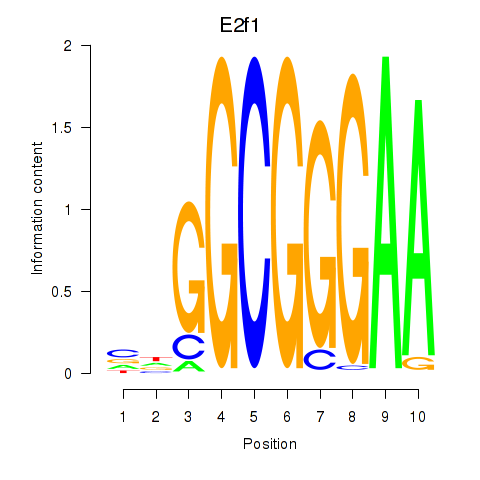
Transcription factors associated with E2f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f1 | ENSMUSG00000027490.11 | E2f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f1 | mm10_v2_chr2_-_154569720_154569799 | 0.86 | 6.2e-17 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.7 | 62.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 19.7 | 59.0 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 17.8 | 71.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 17.1 | 34.2 | GO:0072180 | mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 15.7 | 188.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 14.9 | 74.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 14.2 | 70.8 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 14.1 | 42.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 12.8 | 89.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 12.0 | 35.9 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 11.8 | 23.5 | GO:0007418 | ventral midline development(GO:0007418) |
| 11.2 | 33.5 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 10.8 | 43.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 10.5 | 157.3 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 10.2 | 51.2 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 10.0 | 30.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 9.9 | 29.8 | GO:0097402 | neuroblast migration(GO:0097402) |
| 9.6 | 67.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 9.2 | 45.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 9.1 | 36.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 8.9 | 26.7 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 8.8 | 70.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 8.7 | 35.0 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 8.7 | 26.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 8.1 | 24.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 8.0 | 24.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 7.6 | 22.8 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 7.5 | 30.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 7.4 | 155.2 | GO:0007530 | sex determination(GO:0007530) |
| 7.3 | 249.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 7.2 | 21.7 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 7.2 | 21.6 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 7.0 | 48.7 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 6.8 | 20.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 6.7 | 20.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 6.6 | 124.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 6.4 | 19.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 6.4 | 32.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 6.4 | 12.8 | GO:0033864 | positive regulation of NAD(P)H oxidase activity(GO:0033864) |
| 6.2 | 6.2 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 6.0 | 30.1 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 6.0 | 108.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 5.9 | 5.9 | GO:0072069 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 5.9 | 29.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 5.8 | 11.6 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 5.8 | 52.2 | GO:0033504 | floor plate development(GO:0033504) |
| 5.7 | 85.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 5.7 | 17.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 5.6 | 16.9 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 5.6 | 16.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 5.6 | 5.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 5.4 | 21.7 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 5.2 | 36.7 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 5.2 | 20.8 | GO:0003360 | brainstem development(GO:0003360) |
| 5.2 | 25.9 | GO:0015705 | iodide transport(GO:0015705) |
| 5.1 | 45.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 5.0 | 20.1 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 5.0 | 19.8 | GO:0030091 | protein repair(GO:0030091) |
| 4.9 | 34.6 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 4.9 | 14.8 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 4.8 | 100.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 4.7 | 37.9 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 4.7 | 9.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 4.7 | 23.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 4.6 | 13.9 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 4.5 | 8.9 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 4.4 | 13.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 4.4 | 4.4 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 4.4 | 13.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 4.4 | 4.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 4.3 | 17.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 4.2 | 12.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 4.2 | 21.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 4.1 | 29.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 4.1 | 4.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 4.1 | 77.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 4.0 | 35.9 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 4.0 | 11.9 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 3.9 | 11.7 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 3.9 | 11.7 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 3.8 | 7.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 3.8 | 19.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 3.8 | 19.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 3.8 | 7.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 3.8 | 7.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 3.7 | 3.7 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 3.7 | 14.7 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 3.6 | 14.6 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 3.6 | 10.9 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 3.5 | 17.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 3.5 | 10.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 3.3 | 23.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 3.3 | 16.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 3.3 | 3.3 | GO:0072284 | metanephric S-shaped body morphogenesis(GO:0072284) |
| 3.2 | 22.2 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 3.1 | 30.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 3.0 | 18.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 3.0 | 33.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 3.0 | 29.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 2.9 | 5.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 2.9 | 8.7 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 2.9 | 8.6 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 2.9 | 11.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.8 | 16.8 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 2.8 | 11.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 2.7 | 2.7 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 2.7 | 8.0 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 2.6 | 7.9 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 2.6 | 18.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 2.5 | 20.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 2.5 | 15.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 2.5 | 12.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 2.5 | 9.8 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 2.5 | 4.9 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 2.4 | 14.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 2.4 | 12.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 2.4 | 19.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 2.4 | 2.4 | GO:0071824 | protein-DNA complex subunit organization(GO:0071824) |
| 2.4 | 11.8 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 2.4 | 2.4 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 2.3 | 14.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 2.3 | 28.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 2.3 | 11.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 2.3 | 9.3 | GO:0006526 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) arginine biosynthetic process(GO:0006526) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 2.3 | 9.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 2.3 | 4.6 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 2.3 | 6.9 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 2.3 | 50.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 2.3 | 22.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 2.2 | 4.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 2.2 | 24.4 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 2.2 | 30.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 2.2 | 6.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.1 | 8.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 2.1 | 27.2 | GO:0030903 | notochord development(GO:0030903) |
| 2.1 | 2.1 | GO:0045716 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 2.1 | 16.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 2.1 | 37.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 2.1 | 32.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 2.0 | 24.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 2.0 | 4.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.0 | 10.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 2.0 | 19.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 2.0 | 64.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 1.9 | 19.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.9 | 7.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 1.9 | 7.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 1.8 | 7.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 1.8 | 22.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 1.8 | 11.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.8 | 18.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.8 | 5.4 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 1.8 | 21.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.8 | 5.3 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 1.7 | 20.6 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 1.7 | 6.9 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 1.7 | 10.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 1.7 | 5.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.7 | 3.4 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 1.7 | 1.7 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 1.7 | 5.0 | GO:0061743 | motor learning(GO:0061743) |
| 1.7 | 9.9 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 1.7 | 34.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 1.6 | 11.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.6 | 6.5 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 1.6 | 4.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.6 | 12.7 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.6 | 11.0 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 1.6 | 6.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.6 | 3.1 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 1.6 | 21.7 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 1.5 | 1.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.5 | 3.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.5 | 1.5 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 1.5 | 66.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 1.5 | 3.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.5 | 4.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.4 | 18.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.4 | 5.7 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 1.4 | 4.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 1.4 | 26.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 1.4 | 5.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.4 | 4.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.4 | 23.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 1.4 | 5.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 1.4 | 5.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.4 | 12.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 1.4 | 16.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 1.4 | 5.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 1.4 | 2.7 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 1.4 | 11.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 1.4 | 13.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 1.4 | 10.8 | GO:0001842 | neural fold formation(GO:0001842) |
| 1.3 | 55.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 1.3 | 18.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.3 | 4.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.3 | 10.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 1.3 | 4.0 | GO:2000319 | negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 1.3 | 4.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 1.3 | 5.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.3 | 3.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 1.3 | 11.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 1.3 | 28.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 1.2 | 11.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 1.2 | 3.7 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 1.2 | 2.5 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) phenotypic switching(GO:0036166) |
| 1.2 | 8.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 1.2 | 4.9 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 1.2 | 6.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 1.2 | 9.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.2 | 12.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 1.2 | 23.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 1.2 | 44.6 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 1.2 | 26.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 1.2 | 3.5 | GO:1904154 | protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.2 | 2.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 1.1 | 10.3 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 1.1 | 2.3 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 1.1 | 3.4 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 1.1 | 13.6 | GO:0046605 | regulation of centrosome cycle(GO:0046605) |
| 1.1 | 3.2 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.1 | 1.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 1.1 | 33.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 1.1 | 3.2 | GO:2000047 | regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 1.1 | 5.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.1 | 22.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 1.1 | 6.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 1.0 | 3.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.0 | 5.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 1.0 | 3.1 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 1.0 | 25.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 1.0 | 6.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 1.0 | 6.0 | GO:0097421 | liver regeneration(GO:0097421) |
| 1.0 | 2.0 | GO:0035989 | tendon development(GO:0035989) |
| 1.0 | 5.9 | GO:0035878 | nail development(GO:0035878) |
| 1.0 | 10.7 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 1.0 | 2.9 | GO:0071224 | positive regulation of immature T cell proliferation(GO:0033091) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) response to interleukin-12(GO:0070671) cellular response to peptidoglycan(GO:0071224) |
| 1.0 | 7.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 1.0 | 3.8 | GO:0010713 | negative regulation of collagen metabolic process(GO:0010713) negative regulation of collagen biosynthetic process(GO:0032966) |
| 1.0 | 5.7 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 1.0 | 2.9 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.9 | 4.7 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.9 | 8.5 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.9 | 0.9 | GO:0042368 | negative regulation of vitamin D biosynthetic process(GO:0010957) vitamin D biosynthetic process(GO:0042368) negative regulation of vitamin metabolic process(GO:0046137) regulation of vitamin D biosynthetic process(GO:0060556) |
| 0.9 | 7.4 | GO:0060430 | lung saccule development(GO:0060430) |
| 0.9 | 3.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.9 | 10.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 22.8 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.9 | 32.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.9 | 3.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.9 | 15.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.8 | 3.4 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.8 | 6.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.8 | 2.5 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.8 | 3.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.8 | 2.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.8 | 4.8 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.8 | 12.7 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.8 | 2.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.8 | 2.3 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.8 | 10.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.7 | 3.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.7 | 48.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.7 | 3.6 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.7 | 2.2 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.7 | 0.7 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.7 | 1.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.7 | 2.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.7 | 0.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.7 | 2.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.7 | 3.5 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.7 | 9.6 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.7 | 3.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 17.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.7 | 12.7 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.7 | 1.3 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.7 | 2.6 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.6 | 3.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.6 | 12.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.6 | 0.6 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.6 | 1.9 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.6 | 10.6 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.6 | 12.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.6 | 3.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.6 | 17.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.6 | 5.9 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.6 | 5.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.6 | 1.7 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.6 | 2.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.6 | 9.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.6 | 24.9 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.6 | 7.4 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.6 | 5.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.5 | 6.0 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.5 | 8.0 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.5 | 7.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.5 | 10.0 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.5 | 3.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.5 | 3.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.5 | 19.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.5 | 11.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.5 | 4.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.5 | 6.9 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.5 | 1.9 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.5 | 11.5 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.5 | 1.9 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.5 | 33.3 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.5 | 8.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.5 | 0.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.5 | 9.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 10.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.5 | 0.9 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.5 | 4.5 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 2.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.5 | 1.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.5 | 5.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.4 | 4.0 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.4 | 3.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.4 | 14.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.4 | 21.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.4 | 2.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.4 | 5.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.4 | 6.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.4 | 2.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.4 | 15.4 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.4 | 3.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.4 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.4 | 0.8 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.4 | 2.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 3.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.4 | 4.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.4 | 8.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.4 | 1.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.4 | 0.4 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.4 | 1.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 3.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.4 | 1.9 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.4 | 4.8 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.4 | 1.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.4 | 2.6 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) specification of loop of Henle identity(GO:0072086) |
| 0.4 | 5.8 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.4 | 3.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.3 | 8.2 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.3 | 11.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 1.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 4.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) spermidine biosynthetic process(GO:0008295) |
| 0.3 | 1.3 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.3 | 11.1 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.3 | 1.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 2.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.3 | 3.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 2.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 1.7 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.3 | 3.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 0.8 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 | 2.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 23.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.3 | 1.6 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.3 | 1.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.3 | 1.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.3 | 2.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.3 | 2.1 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.3 | 7.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.3 | 2.3 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.3 | 2.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.3 | 2.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 2.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 1.2 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.2 | 0.5 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 0.5 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.2 | 7.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 3.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 15.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.2 | 1.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 9.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 5.6 | GO:0007492 | endoderm development(GO:0007492) |
| 0.2 | 0.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 3.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 34.4 | GO:0006260 | DNA replication(GO:0006260) |
| 0.2 | 2.8 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.2 | 0.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 0.4 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 1.0 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.2 | 3.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 4.3 | GO:0048599 | oocyte development(GO:0048599) |
| 0.2 | 6.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 3.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 0.8 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.2 | 1.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 2.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.4 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.2 | 1.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 2.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.4 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 4.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 9.0 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.2 | 6.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 12.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.2 | 1.9 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 6.2 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.2 | 19.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 1.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.5 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 1.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 7.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.4 | GO:1904017 | positive regulation of female receptivity(GO:0045925) response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 1.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 3.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 3.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.9 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.5 | GO:0035561 | regulation of chromatin binding(GO:0035561) positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 4.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.2 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 4.5 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.7 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 4.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.0 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.1 | 1.0 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.1 | 0.8 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.1 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 2.7 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 0.9 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.1 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 2.1 | GO:0007588 | excretion(GO:0007588) |
| 0.1 | 0.8 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.1 | 2.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.8 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 4.6 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.1 | 2.7 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.1 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 3.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.0 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 1.0 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.1 | 2.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 6.4 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 5.6 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 5.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.3 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.6 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 1.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 3.7 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.3 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.7 | GO:0001819 | positive regulation of cytokine production(GO:0001819) |
| 0.0 | 0.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.6 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.9 | 68.7 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 18.2 | 90.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 14.8 | 177.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 12.4 | 37.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 12.3 | 37.0 | GO:0035101 | FACT complex(GO:0035101) |
| 11.6 | 34.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 10.5 | 136.5 | GO:0042555 | MCM complex(GO:0042555) |
| 10.3 | 61.7 | GO:0098536 | deuterosome(GO:0098536) |
| 9.7 | 97.5 | GO:0000796 | condensin complex(GO:0000796) |
| 9.4 | 47.1 | GO:0031523 | Myb complex(GO:0031523) |
| 9.1 | 27.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 8.8 | 35.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 8.6 | 25.9 | GO:0000801 | central element(GO:0000801) |
| 6.9 | 20.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 6.7 | 20.2 | GO:0071914 | prominosome(GO:0071914) |
| 6.7 | 33.5 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 6.5 | 19.4 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 6.0 | 36.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 5.5 | 21.9 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 5.0 | 5.0 | GO:0044301 | climbing fiber(GO:0044301) |
| 4.6 | 23.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 4.4 | 145.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 4.4 | 26.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 4.2 | 21.0 | GO:0001740 | Barr body(GO:0001740) |
| 4.2 | 16.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 4.1 | 24.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 3.9 | 11.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 3.9 | 35.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 3.8 | 11.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 3.6 | 17.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 3.4 | 24.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 3.4 | 215.4 | GO:0005657 | replication fork(GO:0005657) |
| 3.3 | 16.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 3.2 | 64.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 3.2 | 12.7 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 3.0 | 18.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.8 | 17.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 2.8 | 5.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 2.6 | 7.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 2.6 | 44.4 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 2.6 | 12.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 2.6 | 10.2 | GO:0008623 | CHRAC(GO:0008623) |
| 2.5 | 12.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 2.4 | 33.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 2.3 | 7.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 2.3 | 11.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 2.2 | 45.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.2 | 6.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 2.1 | 81.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 2.1 | 6.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 2.1 | 35.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 2.0 | 18.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 2.0 | 15.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.9 | 7.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.9 | 5.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.9 | 18.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 1.8 | 7.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.8 | 19.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.8 | 12.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.8 | 10.6 | GO:0000938 | GARP complex(GO:0000938) |
| 1.7 | 5.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.7 | 13.8 | GO:0031415 | NatA complex(GO:0031415) |
| 1.6 | 9.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.6 | 52.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 1.6 | 11.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.5 | 6.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 1.5 | 4.6 | GO:1990075 | stereocilia ankle link complex(GO:0002142) periciliary membrane compartment(GO:1990075) |
| 1.5 | 35.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 1.5 | 10.7 | GO:0097422 | tubular endosome(GO:0097422) |
| 1.5 | 13.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 1.5 | 4.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.5 | 14.6 | GO:0030057 | desmosome(GO:0030057) |
| 1.4 | 5.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 1.4 | 8.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 1.4 | 39.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 1.3 | 15.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 1.3 | 2.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 1.3 | 60.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 1.2 | 20.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.2 | 6.0 | GO:0072487 | MSL complex(GO:0072487) |
| 1.2 | 8.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.2 | 15.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 1.2 | 8.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.2 | 11.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 1.1 | 8.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.1 | 113.4 | GO:0005814 | centriole(GO:0005814) |
| 1.1 | 5.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 1.1 | 17.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 1.1 | 2.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.0 | 13.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.0 | 7.3 | GO:0002177 | manchette(GO:0002177) |
| 1.0 | 20.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 1.0 | 4.9 | GO:0001652 | granular component(GO:0001652) |
| 0.9 | 10.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.9 | 142.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.9 | 6.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.9 | 12.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.8 | 5.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.8 | 39.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.8 | 3.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.8 | 2.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.8 | 2.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.8 | 8.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.8 | 4.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.8 | 2.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.8 | 10.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.8 | 0.8 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.7 | 2.9 | GO:0032021 | NELF complex(GO:0032021) |
| 0.7 | 3.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.7 | 3.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 2.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.7 | 2.8 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.7 | 6.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.7 | 40.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.7 | 10.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 4.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.6 | 3.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.6 | 1.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.6 | 41.2 | GO:0005844 | polysome(GO:0005844) |
| 0.6 | 22.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.6 | 5.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 27.1 | GO:0043034 | costamere(GO:0043034) |
| 0.6 | 3.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.6 | 12.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.6 | 3.9 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.5 | 213.1 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.5 | 21.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 24.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 3.0 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.5 | 4.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 4.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.5 | 1.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.5 | 1.8 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.4 | 11.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 5.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 2.8 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.4 | 39.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.4 | 5.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.4 | 1.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 2.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 11.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.4 | 4.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.4 | 3.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 8.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.3 | 2.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 3.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 3.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 2.6 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 1.9 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 5.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 4.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 57.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 2.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 2.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 58.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.3 | 10.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 8.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 1.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 1.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 10.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 46.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 40.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 11.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 2.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 2.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 28.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 3.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 16.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 7.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 2.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 20.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 5.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 5.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 10.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 17.3 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 9.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 9.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 5.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 685.3 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 4.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) M band(GO:0031430) |
| 0.1 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 2.0 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 6.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.1 | 175.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 32.7 | 130.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 18.2 | 90.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 14.6 | 58.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 13.8 | 41.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 11.8 | 70.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 11.5 | 80.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 11.0 | 32.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 8.8 | 44.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 8.6 | 34.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 8.3 | 25.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 6.9 | 20.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 6.8 | 20.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 6.5 | 25.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 6.4 | 38.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 6.4 | 12.7 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 6.4 | 31.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 5.7 | 45.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 5.5 | 16.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 5.3 | 16.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 4.9 | 34.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 4.9 | 29.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 4.8 | 52.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 4.8 | 19.0 | GO:0042806 | fucose binding(GO:0042806) |
| 4.7 | 42.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 4.5 | 53.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 4.4 | 70.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 4.4 | 163.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 4.3 | 12.9 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 4.2 | 21.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 4.1 | 4.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 3.5 | 3.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 3.5 | 20.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 3.4 | 13.5 | GO:0005113 | patched binding(GO:0005113) |
| 3.3 | 3.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 3.3 | 13.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 3.2 | 19.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 3.2 | 12.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 3.1 | 9.3 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 3.0 | 17.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 3.0 | 14.9 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 3.0 | 11.8 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 2.9 | 14.7 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 2.9 | 23.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.9 | 94.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 2.9 | 11.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 2.9 | 22.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 2.8 | 17.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 2.8 | 8.4 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 2.7 | 8.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 2.7 | 8.0 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 2.5 | 27.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 2.5 | 7.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 2.4 | 19.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.4 | 35.9 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 2.3 | 13.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 2.3 | 112.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 2.3 | 9.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 2.2 | 8.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 2.1 | 6.4 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 2.0 | 6.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 2.0 | 18.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.0 | 9.9 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 1.9 | 11.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.9 | 15.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.9 | 45.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.9 | 7.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.8 | 7.4 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 1.8 | 34.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 1.8 | 17.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.7 | 8.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.7 | 8.7 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 1.7 | 12.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.7 | 12.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 1.7 | 61.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 1.7 | 8.5 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.7 | 67.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.7 | 23.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 1.7 | 1.7 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.6 | 6.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 1.6 | 8.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.6 | 11.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.6 | 9.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.6 | 26.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.5 | 16.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.5 | 6.0 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.5 | 31.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.5 | 5.9 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 1.5 | 4.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 1.4 | 310.0 | GO:0042393 | histone binding(GO:0042393) |
| 1.3 | 26.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 1.3 | 5.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.3 | 4.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 1.3 | 3.9 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 1.3 | 11.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.3 | 7.7 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 1.3 | 7.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 1.3 | 5.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.3 | 8.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.2 | 1.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.2 | 124.0 | GO:0004386 | helicase activity(GO:0004386) |
| 1.2 | 5.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 1.2 | 5.9 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 1.2 | 3.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 1.2 | 9.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 1.2 | 5.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 1.1 | 11.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 1.1 | 27.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.1 | 14.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 1.1 | 6.7 | GO:0089720 | caspase binding(GO:0089720) |
| 1.1 | 6.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.1 | 5.5 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 1.1 | 19.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 1.1 | 4.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.1 | 13.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 1.0 | 62.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 1.0 | 3.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.0 | 8.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.0 | 3.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.0 | 8.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.0 | 12.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.9 | 7.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.9 | 7.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.9 | 24.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.9 | 8.3 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.9 | 10.7 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.9 | 13.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.9 | 18.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.9 | 39.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.8 | 30.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.8 | 1.6 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.8 | 2.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.8 | 11.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.8 | 4.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 8.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.8 | 14.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.8 | 11.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.8 | 28.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.8 | 11.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.8 | 6.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 2.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.8 | 119.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.8 | 2.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.7 | 5.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.7 | 13.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.7 | 5.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.7 | 9.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.7 | 15.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.7 | 7.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 6.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.7 | 12.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.7 | 14.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.7 | 27.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.7 | 2.6 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.6 | 7.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.6 | 5.7 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.6 | 3.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.6 | 2.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.6 | 14.1 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.6 | 4.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.6 | 24.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.6 | 17.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 11.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 16.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 4.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.6 | 5.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 7.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.5 | 2.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.5 | 9.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.5 | 1.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.5 | 4.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.5 | 9.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.5 | 6.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 1.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 3.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.5 | 4.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.4 | 12.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.4 | 23.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.4 | 8.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 11.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 1.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.4 | 26.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.4 | 6.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.4 | 2.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.4 | 11.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 5.0 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.4 | 2.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 2.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 70.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.4 | 1.1 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.3 | 2.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 1.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 1.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 1.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.3 | 12.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 6.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 14.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.3 | 1.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 3.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.3 | 0.6 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.3 | 5.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.3 | 1.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 14.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.3 | 16.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 65.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.3 | 1.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 11.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.2 | 8.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 1.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 8.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 6.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 8.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.2 | 2.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 4.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.2 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 1.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.6 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.2 | 3.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 16.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 5.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 1.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 3.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 2.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 10.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 11.1 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 9.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.8 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.7 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 8.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.5 | GO:0004385 | guanylate kinase activity(GO:0004385) nucleotide kinase activity(GO:0019201) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 42.8 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 14.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 3.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 13.8 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 98.3 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 5.3 | 243.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 4.8 | 19.4 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 4.0 | 321.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 3.9 | 189.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 3.8 | 209.6 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 3.0 | 94.5 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 2.0 | 9.8 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 1.7 | 12.1 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.6 | 6.5 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 1.6 | 54.7 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 1.5 | 44.9 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 1.5 | 14.9 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 1.4 | 84.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 1.2 | 8.5 | PID_MYC_PATHWAY | C-MYC pathway |
| 1.2 | 14.6 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 1.1 | 10.0 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.1 | 56.2 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 1.0 | 12.7 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 77.7 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.9 | 31.8 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.8 | 81.2 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.8 | 24.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.8 | 15.3 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.7 | 23.1 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.7 | 28.6 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.6 | 29.8 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.6 | 8.4 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.6 | 25.0 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.6 | 16.9 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.6 | 13.8 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.5 | 11.3 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.5 | 18.1 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.5 | 19.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.5 | 15.0 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.5 | 54.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 9.6 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.4 | 14.4 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.4 | 19.8 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.4 | 9.0 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 20.1 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.3 | 5.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 3.4 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.3 | 11.2 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 5.2 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.3 | 5.2 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.2 | 3.2 | PID_ATM_PATHWAY | ATM pathway |
| 0.2 | 9.2 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 4.9 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 8.4 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.6 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.2 | 3.3 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 2.6 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 4.3 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 2.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.9 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.1 | 1.0 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.7 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 0.7 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 1.9 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.1 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.6 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.3 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 5.0 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.1 | 162.4 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 8.8 | 158.6 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 8.2 | 139.3 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 7.1 | 92.6 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 6.1 | 24.4 | REACTOME_LAGGING_STRAND_SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 5.1 | 10.2 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 5.0 | 35.3 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 4.6 | 252.2 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 4.6 | 36.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 4.0 | 44.1 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 3.9 | 90.5 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 3.9 | 89.8 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 3.7 | 33.6 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 3.4 | 161.0 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 3.0 | 26.8 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 2.9 | 14.6 | REACTOME_ACTIVATED_POINT_MUTANTS_OF_FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 2.8 | 19.4 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 2.3 | 32.4 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 2.3 | 38.7 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 2.2 | 17.8 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 2.2 | 53.3 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 2.1 | 42.5 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 2.0 | 6.1 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 2.0 | 28.4 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.8 | 21.8 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 1.7 | 19.9 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.4 | 38.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 1.4 | 11.3 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 1.4 | 23.9 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.3 | 14.7 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 1.3 | 20.3 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.2 | 3.7 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 1.2 | 14.6 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.2 | 23.2 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 1.1 | 34.6 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.1 | 30.8 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 1.0 | 11.4 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.0 | 37.2 | REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 1.0 | 18.1 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.9 | 26.3 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.9 | 66.9 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.9 | 24.1 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.9 | 6.2 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.9 | 8.8 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.9 | 19.0 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.9 | 7.7 | REACTOME_MITOTIC_G2_G2_M_PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.8 | 63.9 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.8 | 20.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.8 | 6.9 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.8 | 3.8 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.8 | 3.0 | REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.7 | 11.9 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.7 | 12.2 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.7 | 15.4 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.6 | 23.6 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.5 | 21.8 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 13.6 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 3.6 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.5 | 6.4 | REACTOME_REGULATION_OF_APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.5 | 3.4 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.5 | 12.1 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.5 | 24.2 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.5 | 0.9 | REACTOME_ABORTIVE_ELONGATION_OF_HIV1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.4 | 5.9 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.4 | 11.5 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.4 | 34.1 | REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.4 | 44.4 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.4 | 3.9 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.4 | 8.5 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 7.1 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 26.1 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 3.7 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 6.7 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 21.7 | REACTOME_ANTIGEN_PROCESSING_CROSS_PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.3 | 7.1 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 3.8 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 54.7 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 17.9 | REACTOME_APOPTOTIC_EXECUTION_PHASE | Genes involved in Apoptotic execution phase |
| 0.3 | 5.4 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 7.3 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.2 | 2.9 | REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.2 | 2.6 | REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.2 | 6.9 | REACTOME_INTERFERON_GAMMA_SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 2.8 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 15.4 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 2.6 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 5.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 13.8 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.3 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.1 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.8 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 0.8 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.8 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.8 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 7.4 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.2 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.9 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 0.5 | REACTOME_RNA_POL_I_TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 1.1 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 3.3 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 3.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.4 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 5.6 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.4 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 3.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.8 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.1 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.6 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.6 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |