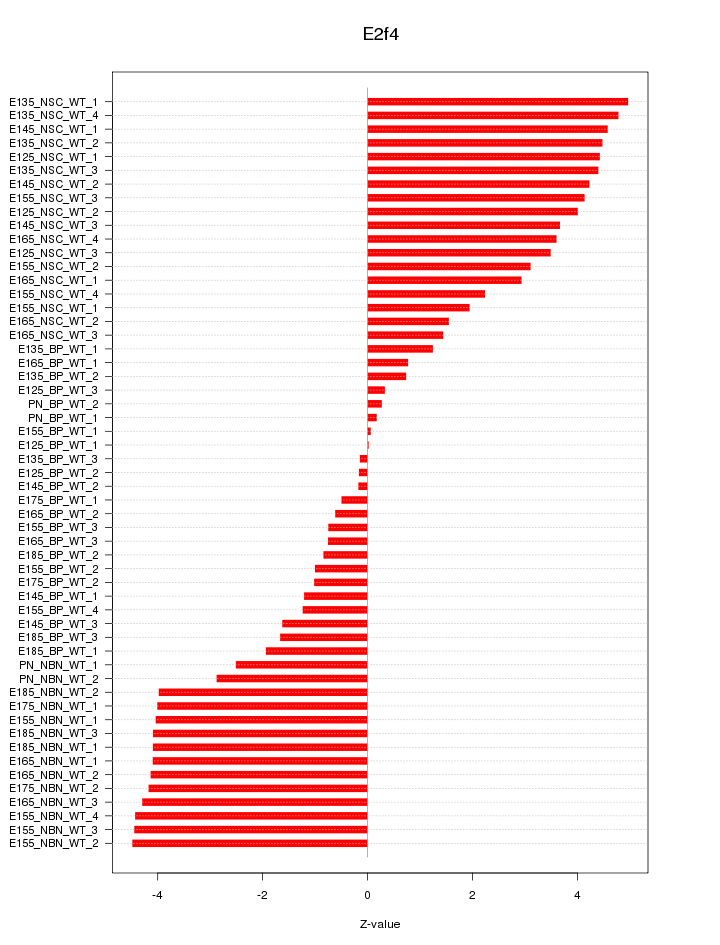
Motif ID: E2f4
Z-value: 2.997
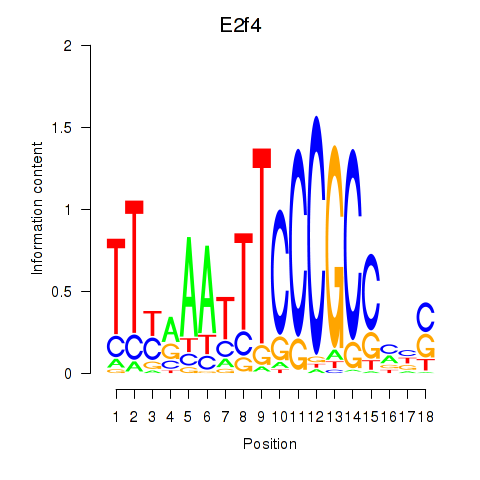
Transcription factors associated with E2f4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f4 | ENSMUSG00000014859.8 | E2f4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | mm10_v2_chr8_+_105297663_105297742 | 0.91 | 5.6e-22 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.1 | 68.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 15.7 | 47.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 10.5 | 31.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 9.7 | 29.2 | GO:0003219 | atrioventricular node development(GO:0003162) cardiac right ventricle formation(GO:0003219) |
| 8.7 | 26.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 6.5 | 19.4 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 5.6 | 16.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 5.1 | 72.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 3.9 | 35.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 3.3 | 22.8 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 2.9 | 72.8 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 2.7 | 16.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 2.6 | 23.0 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 2.4 | 12.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 2.4 | 7.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 2.2 | 33.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 2.2 | 17.2 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 2.1 | 12.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 2.0 | 13.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 2.0 | 73.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 1.9 | 9.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.9 | 46.0 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 1.9 | 51.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 1.9 | 13.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.8 | 3.6 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.6 | 4.9 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 1.5 | 83.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 1.4 | 11.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.2 | 3.7 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 1.2 | 18.0 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 1.2 | 4.8 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 1.2 | 3.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.2 | 6.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.1 | 13.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.1 | 5.4 | GO:1902995 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.1 | 23.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 1.0 | 4.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 1.0 | 34.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 1.0 | 3.9 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.9 | 9.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.9 | 3.4 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 | 3.2 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.8 | 7.8 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.8 | 36.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.7 | 29.9 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.7 | 8.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.6 | 2.6 | GO:0043987 | histone-serine phosphorylation(GO:0035404) histone H3-S10 phosphorylation(GO:0043987) |
| 0.6 | 13.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.6 | 8.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.6 | 18.8 | GO:0007129 | synapsis(GO:0007129) |
| 0.6 | 1.8 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.5 | 3.8 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.5 | 26.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.5 | 10.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.5 | 7.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.5 | 3.7 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.4 | 36.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.4 | 2.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.4 | 1.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 1.6 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.4 | 5.3 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.4 | 2.4 | GO:0035878 | nail development(GO:0035878) |
| 0.4 | 2.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.4 | 1.9 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 5.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 1.0 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 | 22.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 3.4 | GO:0033211 | leptin-mediated signaling pathway(GO:0033210) adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 2.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.5 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 0.6 | GO:1905077 | regulation of histone ubiquitination(GO:0033182) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.2 | 2.0 | GO:0042754 | histone H3-K9 dimethylation(GO:0036123) negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 4.4 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.2 | 0.9 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.2 | 7.3 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 3.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 54.5 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 10.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 | 4.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 1.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0051608 | histamine transport(GO:0051608) |
| 0.1 | 2.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 5.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 1.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 2.1 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 1.3 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 11.9 | GO:0007049 | cell cycle(GO:0007049) |
| 0.0 | 7.8 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 73.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 6.7 | 47.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 6.0 | 29.9 | GO:0031523 | Myb complex(GO:0031523) |
| 5.8 | 29.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 5.3 | 31.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 4.3 | 13.0 | GO:0035101 | FACT complex(GO:0035101) |
| 3.7 | 26.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 3.2 | 16.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 2.8 | 34.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 2.8 | 28.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 2.6 | 23.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 2.1 | 41.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.6 | 11.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 1.4 | 16.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.1 | 72.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.9 | 65.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.7 | 7.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 18.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.6 | 67.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.6 | 41.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.6 | 4.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.6 | 3.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 41.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.4 | 7.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 2.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 2.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 2.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 2.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 23.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.4 | 2.5 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 4.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 18.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 29.8 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 32.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.3 | 12.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.3 | 4.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 5.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 5.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 25.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.2 | 1.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 23.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 12.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 5.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 23.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 27.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 22.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 13.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 9.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 6.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 16.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 86.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 64.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 9.8 | 68.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 7.2 | 36.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 3.7 | 11.2 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 3.7 | 29.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 3.0 | 20.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 2.9 | 26.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.8 | 8.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 2.6 | 26.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 2.4 | 12.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 2.4 | 9.6 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 2.3 | 16.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 2.1 | 23.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.9 | 19.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 1.9 | 16.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 1.8 | 5.4 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.4 | 4.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single guanine insertion binding(GO:0032142) |
| 1.3 | 46.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 1.1 | 3.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 1.0 | 34.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 1.0 | 3.9 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.9 | 13.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.9 | 41.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.9 | 3.4 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.9 | 2.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.8 | 29.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.8 | 23.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.8 | 6.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 7.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 18.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.7 | 2.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.6 | 13.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.6 | 3.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.6 | 35.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.5 | 18.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.5 | 2.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 4.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 3.7 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.4 | 3.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 51.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.3 | 3.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 5.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 1.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 2.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 0.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 1.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 4.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 40.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 9.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 3.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 9.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 7.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 31.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 24.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.3 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 43.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 2.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 2.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 94.9 | PID_ATM_PATHWAY | ATM pathway |
| 1.8 | 19.4 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 1.3 | 18.3 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.1 | 31.1 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 1.1 | 36.8 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 1.0 | 41.2 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.8 | 68.5 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.7 | 28.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.7 | 46.3 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.7 | 68.3 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.5 | 13.0 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.4 | 26.1 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.4 | 12.1 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 2.4 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 6.9 | PID_ERBB1_RECEPTOR_PROXIMAL_PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.3 | 9.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.8 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.2 | 7.9 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.2 | 4.9 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 7.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.5 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 6.1 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 4.4 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.6 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.9 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.6 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 46.0 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 2.9 | 26.1 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 2.6 | 66.1 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 2.5 | 53.3 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.6 | 27.3 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.2 | 12.9 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.1 | 35.0 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.9 | 11.2 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.9 | 98.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.9 | 44.6 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.7 | 66.8 | REACTOME_LATE_PHASE_OF_HIV_LIFE_CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.6 | 4.9 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.5 | 7.0 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 6.9 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.5 | 61.5 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.5 | 22.1 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.3 | 9.6 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 12.1 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 8.4 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.3 | 5.4 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 1.3 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 3.9 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.2 | 3.4 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 3.6 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.7 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.9 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 0.7 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.7 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.3 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |