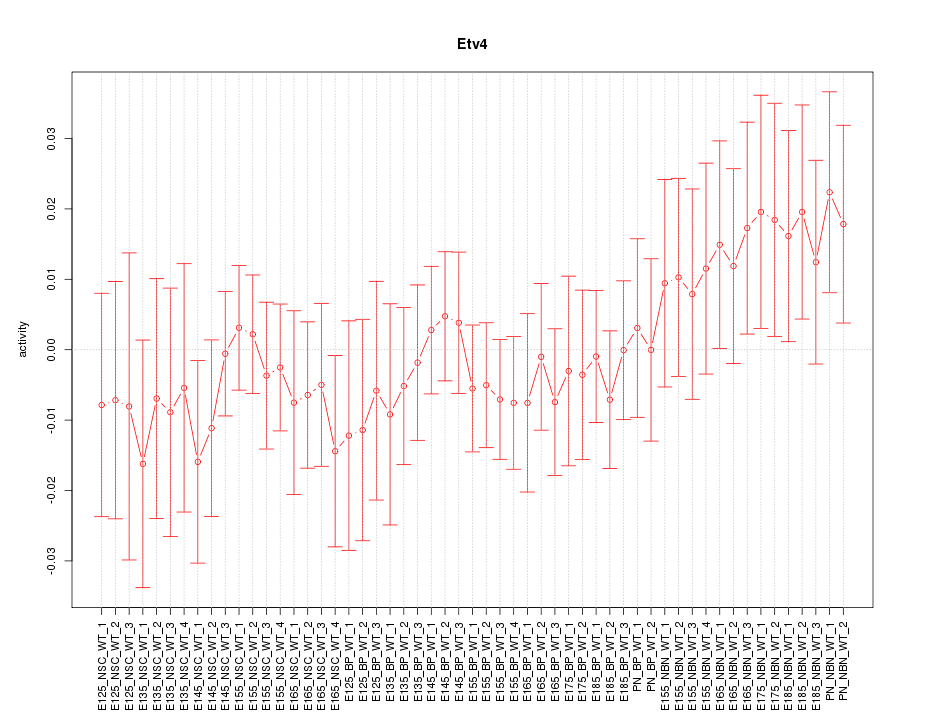
Motif ID: Etv4
Z-value: 0.707

Transcription factors associated with Etv4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Etv4 | ENSMUSG00000017724.8 | Etv4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv4 | mm10_v2_chr11_-_101785252_101785371 | 0.16 | 2.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.0 | 3.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.9 | 3.8 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.9 | 4.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.9 | 5.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.8 | 4.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.8 | 2.4 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.8 | 2.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.7 | 2.8 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.7 | 2.0 | GO:0019046 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) release from viral latency(GO:0019046) |
| 0.7 | 3.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 | 6.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.6 | 3.5 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.5 | 2.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.5 | 4.7 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 1.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.5 | 4.7 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 2.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.5 | 5.5 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.5 | 3.4 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 | 1.5 | GO:0061744 | motor behavior(GO:0061744) |
| 0.5 | 1.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.5 | 1.9 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.5 | 1.4 | GO:1905072 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.5 | 1.4 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 1.8 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.4 | 5.7 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 1.7 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 2.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.4 | 3.3 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.4 | 1.6 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.4 | 2.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 0.7 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.4 | 0.4 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.4 | 1.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.4 | 1.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.4 | 1.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.3 | 1.7 | GO:0010637 | diet induced thermogenesis(GO:0002024) negative regulation of mitochondrial fusion(GO:0010637) |
| 0.3 | 1.7 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 0.7 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 1.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.3 | 1.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 4.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.3 | 1.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.9 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 3.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 7.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.3 | 0.9 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.3 | 2.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 0.6 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.3 | 0.6 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 3.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 0.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 1.1 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.3 | 1.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.8 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.3 | 0.5 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.3 | 1.3 | GO:2000427 | eosinophil chemotaxis(GO:0048245) T cell extravasation(GO:0072683) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 1.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 2.2 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) |
| 0.2 | 2.6 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 1.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 3.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 1.1 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 1.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 0.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 1.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 2.8 | GO:0036296 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.2 | 0.8 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.2 | 0.6 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.2 | 0.6 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.8 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 0.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 0.6 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 0.6 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.2 | 0.8 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.2 | 0.5 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 0.5 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.2 | 1.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 2.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 0.5 | GO:2001180 | regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 0.7 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 2.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.5 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.2 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 1.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 0.8 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.2 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.5 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 1.1 | GO:0021886 | female meiosis I(GO:0007144) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 1.8 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.8 | GO:0090647 | astrocyte activation involved in immune response(GO:0002265) choline transport(GO:0015871) modulation of age-related behavioral decline(GO:0090647) regulation of core promoter binding(GO:1904796) |
| 0.2 | 1.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 | 1.0 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.1 | 0.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 2.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 0.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 2.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 3.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.8 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 1.3 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 1.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.7 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.7 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 3.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.5 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.6 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.5 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 1.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 1.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.7 | GO:0010730 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 6.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 2.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.1 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.1 | 0.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 1.1 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.3 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.6 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 1.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.9 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.8 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.5 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 1.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 1.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.9 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0002692 | negative regulation of cellular extravasation(GO:0002692) |
| 0.1 | 0.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.1 | GO:2000078 | positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.8 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.6 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 0.6 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 1.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.5 | GO:0072015 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 1.3 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 1.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.0 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 4.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.8 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 1.4 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.1 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 0.1 | 0.6 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 2.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 0.7 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.4 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.1 | 2.3 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 1.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.2 | GO:0035247 | peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 3.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 1.9 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 2.1 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.4 | GO:0033683 | transcription-coupled nucleotide-excision repair(GO:0006283) nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 1.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:2000121 | regulation of superoxide dismutase activity(GO:1901668) regulation of removal of superoxide radicals(GO:2000121) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.7 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.6 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 3.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.6 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 3.0 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 1.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.7 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 1.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.5 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 4.0 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 1.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 3.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.1 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:1905216 | positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.4 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of vacuole organization(GO:0044090) positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.3 | 3.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.1 | 5.3 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.9 | 2.6 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.8 | 3.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.7 | 0.7 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.6 | 1.9 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.5 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.5 | 3.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 1.5 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.5 | 3.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 2.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.5 | 3.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.5 | 1.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 2.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 1.3 | GO:0044299 | C-fiber(GO:0044299) |
| 0.4 | 3.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 1.9 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 1.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 1.3 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 1.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 3.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 1.8 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.3 | 0.9 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.3 | 0.5 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.2 | 0.7 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 1.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.8 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.2 | 0.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 5.2 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 7.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 1.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 5.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 2.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.0 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 2.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 12.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 10.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 3.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 2.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 8.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 4.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 3.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0044449 | contractile fiber part(GO:0044449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 1.0 | 2.9 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.9 | 3.4 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.7 | 2.7 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.6 | 2.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 2.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 3.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.6 | 2.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 1.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 1.9 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.5 | 3.2 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.4 | 1.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 4.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.4 | 2.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.4 | 1.2 | GO:0004534 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 6.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 6.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 1.6 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.3 | 1.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 1.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 1.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 1.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 3.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.3 | 1.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.3 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 1.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 3.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 3.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 4.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 4.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 0.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 0.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 2.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 2.9 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.2 | 0.6 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.2 | 0.6 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.2 | 0.6 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 0.6 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.2 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 2.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 3.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 6.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 5.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 0.5 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 0.8 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.2 | 1.3 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 6.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 7.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 3.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.6 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.4 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 2.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 4.3 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.5 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 10.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 2.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 5.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.3 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 3.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 4.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 0.2 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 2.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 5.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 1.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 4.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 1.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 5.6 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.2 | 12.2 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.4 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 5.5 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.1 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.4 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.6 | PID_GMCSF_PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.3 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.4 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.1 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.8 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.6 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.4 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.0 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.8 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.0 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.2 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.5 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.9 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 0.4 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 5.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 5.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.3 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.7 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 3.6 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.4 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 3.6 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.4 | 2.3 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.4 | 7.3 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 4.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.3 | 4.1 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 2.2 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 5.0 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 7.1 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.0 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 2.8 | REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.2 | 3.5 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 4.0 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.0 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 3.0 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.1 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 4.6 | REACTOME_FORMATION_OF_RNA_POL_II_ELONGATION_COMPLEX_ | Genes involved in Formation of RNA Pol II elongation complex |
| 0.1 | 1.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.9 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_MULTIPLE_NUCLEOTIDE_PATCH_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.1 | 7.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.0 | REACTOME_ADVANCED_GLYCOSYLATION_ENDPRODUCT_RECEPTOR_SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.6 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.0 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.3 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 5.3 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 0.9 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.9 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.6 | REACTOME_TRAF6_MEDIATED_INDUCTION_OF_TAK1_COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 0.8 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 1.4 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.3 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.7 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.8 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.4 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.6 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.0 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.3 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.4 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.2 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.8 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 2.2 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.8 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 1.1 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.9 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME_TRNA_AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.4 | REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.6 | REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.5 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |