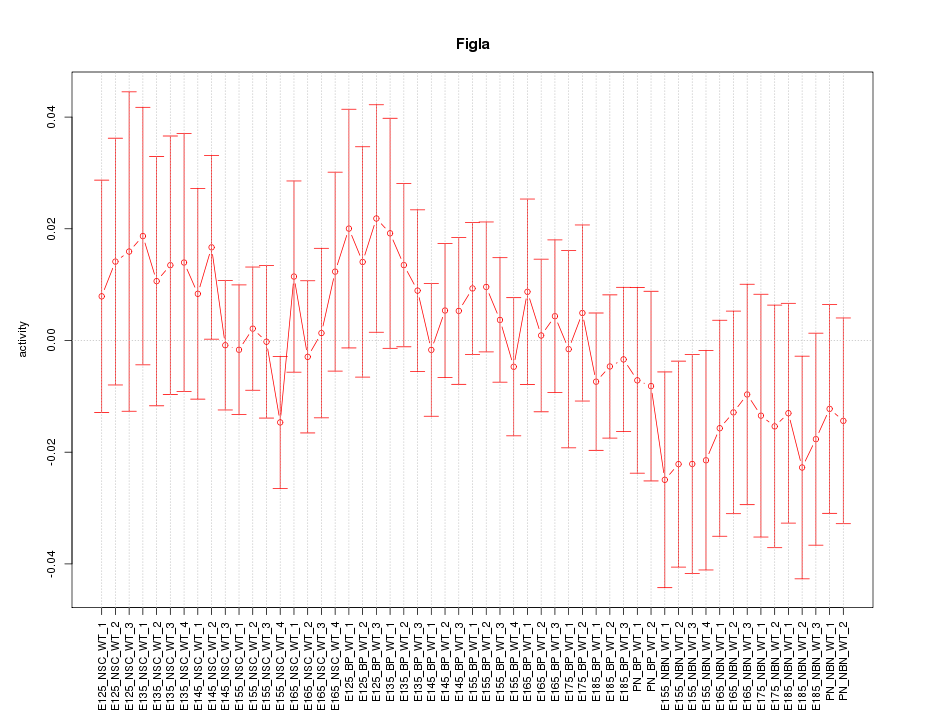
Motif ID: Figla
Z-value: 0.684
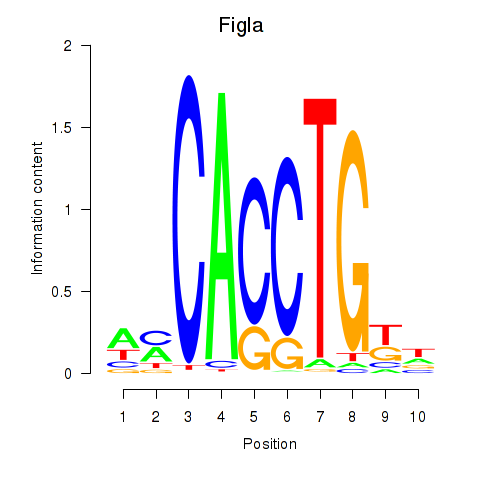
Transcription factors associated with Figla:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Figla | ENSMUSG00000030001.3 | Figla |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 1.8 | 8.8 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 1.7 | 5.2 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.7 | 6.9 | GO:0060032 | notochord regression(GO:0060032) |
| 1.1 | 5.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.1 | 1.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.0 | 3.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 1.0 | 3.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.8 | 2.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.7 | 3.7 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.7 | 2.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.6 | 2.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 | 3.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 6.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.6 | 1.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.6 | 1.8 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.6 | 3.9 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.5 | 2.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 5.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.5 | 2.3 | GO:0019230 | proprioception(GO:0019230) |
| 0.5 | 1.4 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.4 | 1.3 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.4 | 3.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 1.2 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.4 | 5.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.4 | 1.5 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.4 | 1.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.6 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric collecting duct development(GO:0072205) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.3 | 1.0 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.3 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 2.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 3.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 2.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 1.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.2 | 1.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.2 | 1.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 2.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.8 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 0.2 | 2.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 1.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.2 | 0.2 | GO:1901630 | negative regulation of presynaptic membrane organization(GO:1901630) |
| 0.2 | 4.0 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 2.2 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.2 | 1.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 0.5 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.1 | 0.7 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) cellular response to actinomycin D(GO:0072717) |
| 0.1 | 1.3 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 0.6 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.8 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.1 | 1.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.5 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.8 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.5 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 2.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 11.5 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 4.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 1.4 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.1 | 1.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0070886 | positive regulation of NFAT protein import into nucleus(GO:0051533) positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 2.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.5 | GO:0036159 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.9 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.9 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 1.2 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.6 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 3.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 2.1 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 2.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 3.6 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.4 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 2.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0010575 | positive regulation of vascular endothelial growth factor production(GO:0010575) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.3 | 5.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.8 | 3.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.8 | 7.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 6.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 3.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 3.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 10.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 1.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 1.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.3 | 1.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 1.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 2.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 6.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 3.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.3 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.1 | 2.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 3.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 6.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 3.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 4.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.3 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.3 | 5.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.8 | 2.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.7 | 2.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 3.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.4 | 2.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.4 | 3.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.3 | 1.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 2.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 3.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 4.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 0.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.6 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 8.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 6.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 4.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.2 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 5.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 1.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 4.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.8 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 0.4 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 3.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 2.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 1.5 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 1.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 1.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 2.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 1.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.6 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 7.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 8.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 11.0 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.2 | 5.9 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 5.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.1 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.8 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.5 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 3.2 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.3 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.7 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 5.6 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.1 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 2.0 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.3 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.6 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.6 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.2 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.2 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 5.9 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 7.2 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 5.4 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 1.7 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.5 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.0 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.2 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.0 | REACTOME_SIGNALING_BY_FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 2.4 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 7.0 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.5 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.1 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.6 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.2 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 6.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.9 | REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 2.7 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 3.1 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.6 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 2.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME_RNA_POL_II_PRE_TRANSCRIPTION_EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.7 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME_PLATELET_AGGREGATION_PLUG_FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 1.0 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 4.2 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |