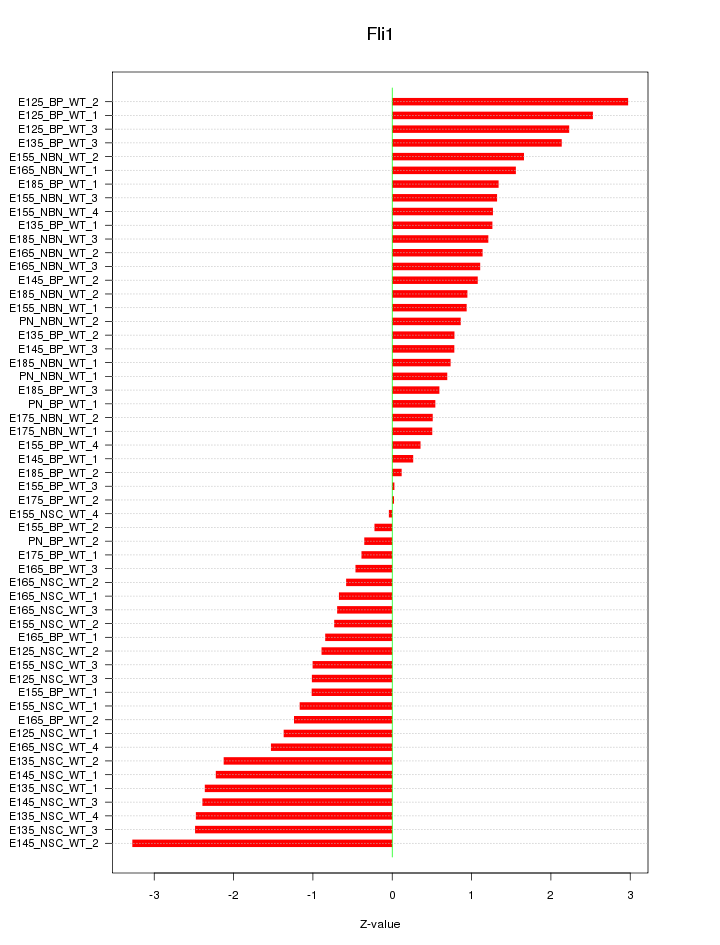
Motif ID: Fli1
Z-value: 1.387
Transcription factors associated with Fli1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Fli1 | ENSMUSG00000016087.7 | Fli1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fli1 | mm10_v2_chr9_-_32541589_32541602 | 0.10 | 4.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0071336 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.9 | 5.8 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.5 | 42.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 1.4 | 5.6 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.4 | 14.9 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 1.3 | 13.9 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.2 | 14.5 | GO:0051823 | radial glia guided migration of Purkinje cell(GO:0021942) regulation of synapse structural plasticity(GO:0051823) |
| 1.1 | 4.3 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.8 | 7.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.8 | 7.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.7 | 2.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.7 | 4.0 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.6 | 1.8 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.5 | 19.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.5 | 2.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.5 | 1.4 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.4 | 55.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.4 | 7.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.4 | 30.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.3 | 2.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 9.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.3 | 4.1 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 1.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 1.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.3 | 1.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 3.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 3.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.2 | 3.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 4.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 1.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 1.0 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.2 | 15.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 2.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 5.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 4.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.2 | 1.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 0.8 | GO:2000774 | positive regulation of Rac protein signal transduction(GO:0035022) positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 1.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 2.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.0 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 3.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.8 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 2.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 2.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.3 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 2.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 2.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 4.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 2.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 3.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 3.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 1.7 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 5.7 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 5.4 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 1.6 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 3.5 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.4 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 55.8 | GO:0044292 | dendrite terminus(GO:0044292) |
| 1.7 | 33.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.3 | 7.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.8 | 2.4 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.8 | 2.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.5 | 4.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 30.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.4 | 7.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 5.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 4.8 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.2 | 0.7 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 14.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 4.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 5.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.1 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 32.2 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 7.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 6.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 4.4 | GO:0045202 | synapse(GO:0045202) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 42.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 3.5 | 63.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 2.5 | 19.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.9 | 5.6 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 1.5 | 6.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.5 | 10.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.5 | 2.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.5 | 31.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.5 | 1.8 | GO:0070891 | peptidoglycan binding(GO:0042834) lipoteichoic acid binding(GO:0070891) |
| 0.4 | 5.8 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.4 | 3.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 4.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 2.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 5.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 13.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.2 | 5.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 4.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 9.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 4.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 4.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 10.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 2.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.5 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 5.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 6.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 13.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 4.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.4 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 3.4 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 6.9 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 7.3 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.2 | 8.3 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.1 | 2.4 | PID_LYMPH_ANGIOGENESIS_PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 4.4 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 3.3 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.0 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.0 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 1.0 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.6 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 33.6 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 6.1 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.4 | 55.8 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.3 | 7.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 5.0 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 9.7 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 4.8 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 1.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 4.5 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 3.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.2 | 2.4 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 6.1 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.7 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.9 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.2 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.0 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.0 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.9 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.8 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.5 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.8 | REACTOME_SHC_MEDIATED_CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 0.4 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 3.1 | REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.1 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 1.5 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |