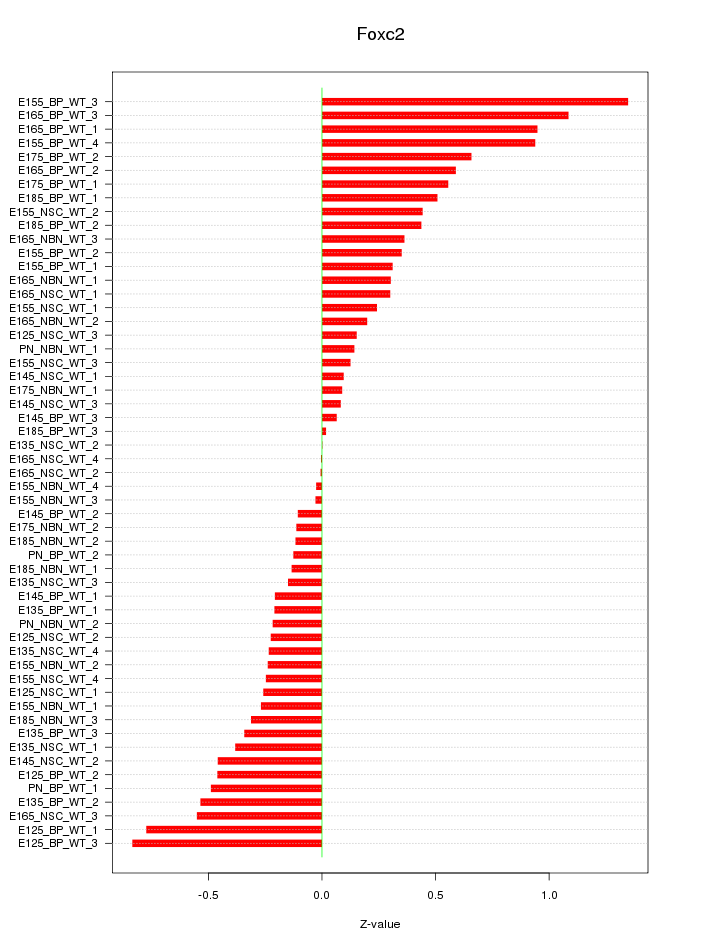
Motif ID: Foxc2
Z-value: 0.442
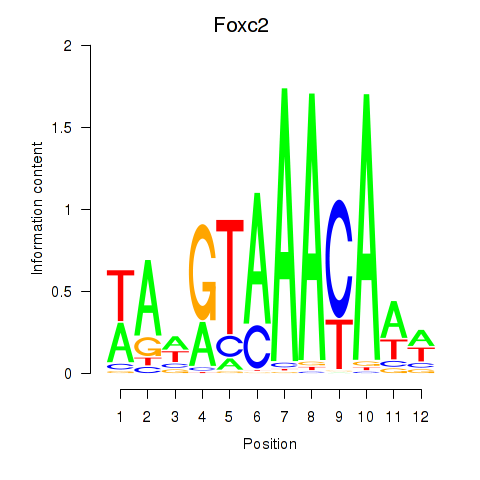
Transcription factors associated with Foxc2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxc2 | ENSMUSG00000046714.6 | Foxc2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc2 | mm10_v2_chr8_+_121116163_121116177 | 0.10 | 4.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.8 | 3.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.3 | 2.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.3 | 0.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 0.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 1.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.2 | 3.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.5 | GO:1902524 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 | 2.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.7 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.8 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.6 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 1.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.6 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.4 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.6 | GO:0072015 | glomerular basement membrane development(GO:0032836) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 2.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.4 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.7 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 1.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 2.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 2.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.7 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 2.4 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.8 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.4 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 2.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |