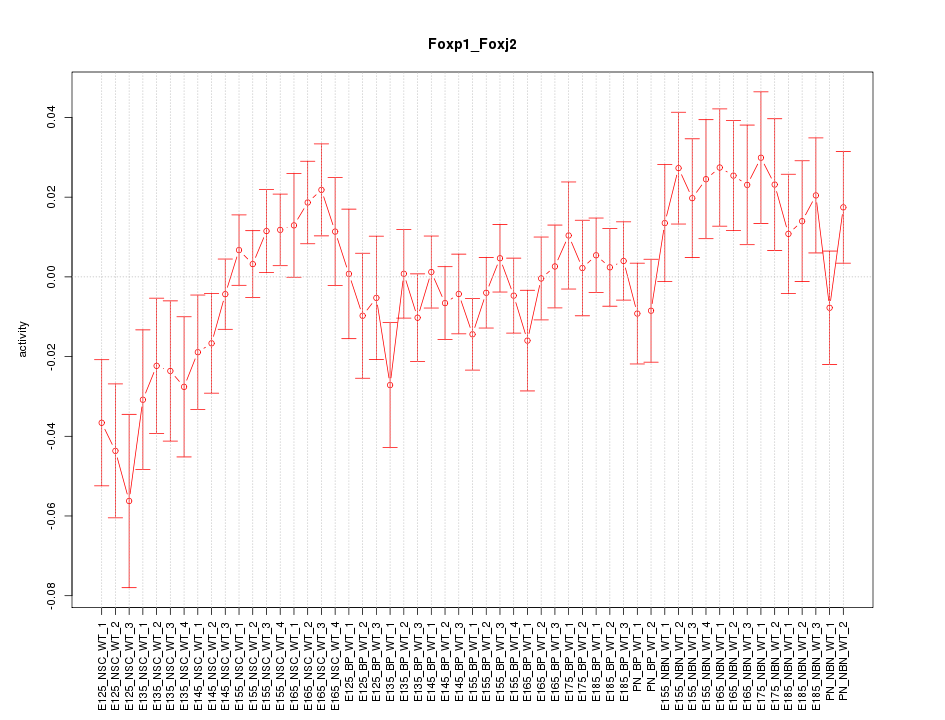
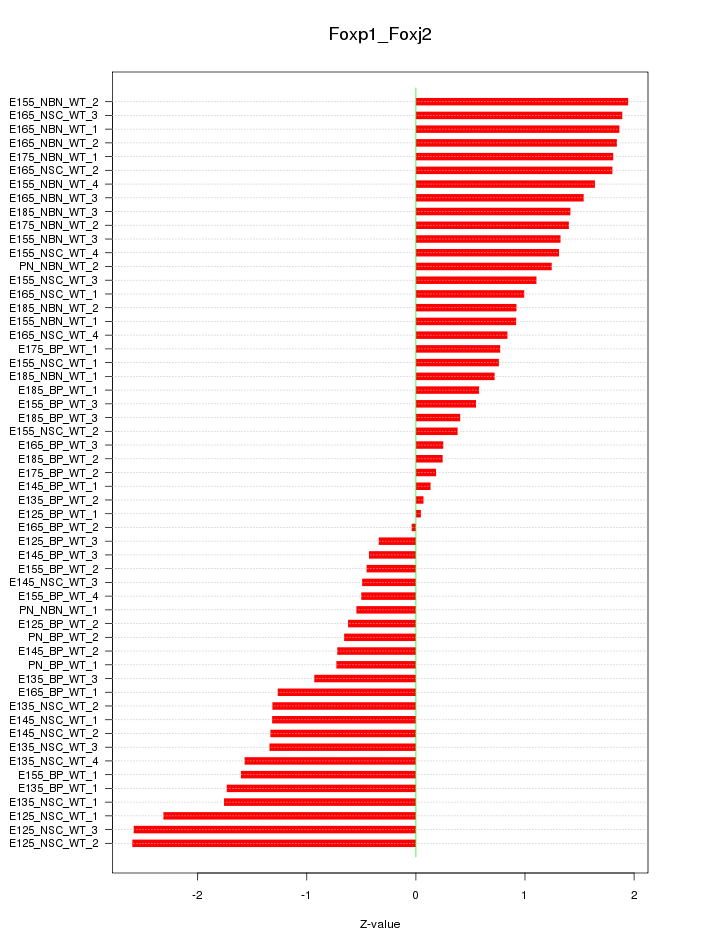
Motif ID: Foxp1_Foxj2
Z-value: 1.242
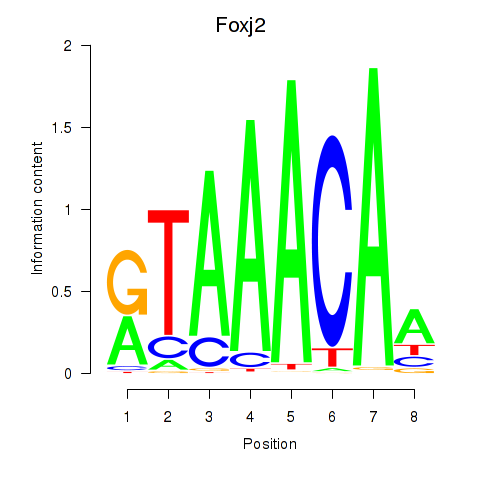
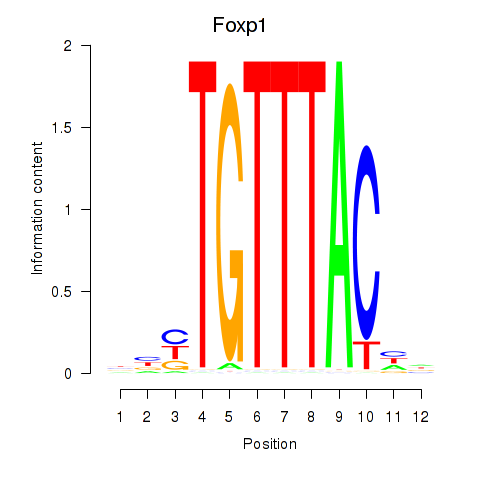
Transcription factors associated with Foxp1_Foxj2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxj2 | ENSMUSG00000003154.9 | Foxj2 |
| Foxp1 | ENSMUSG00000030067.11 | Foxp1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxj2 | mm10_v2_chr6_+_122819888_122819938 | 0.79 | 5.1e-13 | Click! |
| Foxp1 | mm10_v2_chr6_-_99028874_99028942 | 0.60 | 1.5e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 20.0 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 4.0 | 12.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 3.4 | 10.3 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 2.3 | 9.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 2.3 | 6.8 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 2.2 | 6.6 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 2.0 | 26.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 2.0 | 16.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 1.8 | 7.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.8 | 5.4 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 1.7 | 5.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.7 | 6.8 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 1.5 | 7.6 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 1.5 | 4.6 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.5 | 4.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 1.5 | 14.6 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 1.4 | 10.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.4 | 7.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.3 | 1.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 1.3 | 17.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.3 | 3.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.3 | 11.8 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 1.3 | 7.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.2 | 13.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.2 | 3.5 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 1.1 | 7.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.1 | 4.5 | GO:1903294 | axon target recognition(GO:0007412) regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 1.1 | 10.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 7.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 1.1 | 3.3 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 1.1 | 3.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.1 | 7.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 1.1 | 7.5 | GO:0035826 | rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) |
| 1.1 | 3.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 1.1 | 6.4 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 1.0 | 3.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.0 | 5.9 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) optic nerve morphogenesis(GO:0021631) |
| 1.0 | 2.9 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 3.9 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 1.0 | 2.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.9 | 17.7 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.9 | 12.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.9 | 2.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.9 | 7.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.9 | 6.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.9 | 2.6 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.8 | 5.9 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.8 | 5.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.8 | 9.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.8 | 5.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.8 | 5.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.8 | 3.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.8 | 3.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.7 | 2.2 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.7 | 8.1 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.7 | 2.9 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.7 | 5.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.7 | 7.2 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.7 | 3.6 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.7 | 2.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.7 | 2.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.7 | 7.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.7 | 1.4 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.7 | 2.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.7 | 4.9 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.7 | 1.4 | GO:0045922 | negative regulation of fatty acid biosynthetic process(GO:0045717) negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.7 | 7.6 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.7 | 2.7 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.7 | 7.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.7 | 5.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.6 | 1.3 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.6 | 1.9 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 10.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.6 | 10.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.6 | 1.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.6 | 1.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.6 | 1.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.6 | 2.4 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.6 | 4.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.6 | 1.8 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.6 | 4.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.6 | 2.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.6 | 3.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.6 | 4.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.6 | 16.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.6 | 6.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.6 | 5.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.6 | 1.7 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.6 | 1.7 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.6 | 1.7 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.6 | 2.8 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.5 | 4.3 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.5 | 1.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.5 | 2.6 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.5 | 3.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.5 | 2.0 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.5 | 2.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 2.0 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.5 | 1.5 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.5 | 2.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.5 | 1.9 | GO:0044849 | estrous cycle(GO:0044849) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.5 | 2.8 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.5 | 1.9 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.5 | 1.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 2.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 1.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 | 1.8 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 12.7 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.4 | 1.7 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.4 | 3.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.4 | 5.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.4 | 1.7 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.4 | 3.4 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.4 | 1.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 17.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.4 | 1.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.4 | 5.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.4 | 4.8 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.4 | 3.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 2.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.4 | 2.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.4 | 4.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 1.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 1.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 7.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 5.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.3 | 2.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 7.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 1.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.3 | 8.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 1.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 | 1.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.3 | 1.9 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.3 | 1.2 | GO:0050904 | diapedesis(GO:0050904) |
| 0.3 | 4.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 4.6 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.3 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 1.5 | GO:0097646 | positive regulation of protein glycosylation(GO:0060050) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 1.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 2.1 | GO:0090494 | catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.3 | 1.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 3.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.3 | 2.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 2.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 3.4 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.3 | 4.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.3 | 6.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.3 | 0.8 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 0.8 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 1.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 3.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.3 | 0.5 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.3 | 9.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 1.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.3 | 1.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 4.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 3.5 | GO:0098722 | asymmetric stem cell division(GO:0098722) |
| 0.2 | 3.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.2 | 1.0 | GO:2000858 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 3.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.7 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.2 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 0.5 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 1.4 | GO:0046103 | ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.2 | 2.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.7 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 0.7 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 4.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 1.8 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.2 | 1.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.7 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.2 | 2.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.2 | 1.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 2.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.9 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.2 | 2.4 | GO:0042759 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.9 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 1.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 1.3 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.2 | 1.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 4.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 0.8 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 2.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.2 | 2.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 16.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 1.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 3.9 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 2.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.2 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 3.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 1.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 2.7 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.2 | 3.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 7.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 1.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 4.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 0.7 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 2.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 1.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.3 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 3.0 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.2 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 1.0 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 12.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.5 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.2 | 0.3 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.2 | 2.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 0.8 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 3.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.2 | 0.5 | GO:0002638 | negative regulation of immunoglobulin production(GO:0002638) |
| 0.2 | 1.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.2 | 1.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.2 | 1.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.9 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.2 | 1.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 2.7 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.2 | 2.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.2 | 1.8 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.9 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.4 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.1 | 2.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 2.0 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 1.0 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.7 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.7 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.5 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 4.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 2.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 3.1 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 0.5 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.4 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.1 | 2.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.9 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 1.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 3.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 1.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 2.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.8 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.9 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.7 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.1 | 1.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 2.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.5 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.9 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.5 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 2.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 2.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.8 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 0.4 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 1.5 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.9 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.4 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 1.6 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.1 | 5.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.7 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 8.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 10.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 | 2.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 2.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) positive regulation of glucose import(GO:0046326) |
| 0.1 | 2.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.0 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 1.1 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.3 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 1.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.1 | GO:0045359 | positive regulation of chemokine biosynthetic process(GO:0045080) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 1.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 1.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.5 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.5 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 0.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.7 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 1.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.7 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.1 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.5 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.2 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 3.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 4.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.5 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 2.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0014842 | skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 | 0.3 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 1.7 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.3 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.3 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.2 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0043512 | inhibin A complex(GO:0043512) |
| 2.0 | 6.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.9 | 7.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.7 | 7.0 | GO:0031673 | H zone(GO:0031673) |
| 1.5 | 4.4 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.1 | 9.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.0 | 4.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.9 | 4.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.9 | 4.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 15.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.8 | 10.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.7 | 9.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 21.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 3.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.6 | 1.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.6 | 7.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 1.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.6 | 10.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.6 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 4.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.5 | 4.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 2.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 4.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.5 | 1.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.5 | 2.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 1.4 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.4 | 2.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.4 | 1.8 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.4 | 10.0 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 2.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.4 | 2.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.4 | 1.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 3.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 1.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 4.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 5.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 1.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 5.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 2.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 4.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 4.2 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 1.5 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.3 | 0.9 | GO:0044753 | amphisome(GO:0044753) |
| 0.3 | 0.6 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 2.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 10.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.3 | 1.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 2.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 8.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 2.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 2.3 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 1.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 3.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.7 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 5.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 4.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 2.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 3.3 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 0.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 11.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 4.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 3.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 2.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 7.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 1.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.2 | 1.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 2.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 3.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 15.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 2.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 3.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 52.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.2 | 1.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 6.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 7.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 9.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.9 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 6.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 4.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 18.4 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 5.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 3.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 20.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 2.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 1.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 3.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 12.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 13.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.7 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 46.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.3 | GO:0038201 | TOR complex(GO:0038201) |
| 0.1 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 11.6 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 4.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 3.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 3.4 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 2.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 5.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 32.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 3.2 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 2.1 | 6.4 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.9 | 5.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 1.9 | 7.6 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.5 | 5.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.5 | 7.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.5 | 5.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.3 | 4.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 1.3 | 5.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.3 | 7.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.3 | 7.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.2 | 1.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 1.2 | 9.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.1 | 7.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.1 | 4.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.1 | 3.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.1 | 7.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.1 | 11.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.0 | 5.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.0 | 7.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.9 | 10.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.9 | 2.6 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 3.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.8 | 6.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.8 | 2.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.8 | 4.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.8 | 2.4 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.8 | 3.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.8 | 3.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.8 | 4.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 2.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 3.6 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.7 | 7.9 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.7 | 2.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.7 | 5.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 4.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.7 | 2.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 16.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.7 | 2.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.7 | 18.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 4.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 3.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 1.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.6 | 2.4 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.6 | 3.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.6 | 4.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.6 | 3.6 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.6 | 1.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.6 | 2.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.6 | 1.7 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.5 | 3.8 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.5 | 4.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.5 | 1.6 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.5 | 4.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.5 | 2.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 5.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 15.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.5 | 9.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 6.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 2.8 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.5 | 13.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 3.9 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 5.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 1.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.4 | 1.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.4 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 3.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 1.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 1.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 4.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 1.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 2.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 3.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 1.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.3 | 3.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 18.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.3 | 7.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 1.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 2.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 1.9 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 1.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 4.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 2.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.5 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.3 | 2.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 11.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 2.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.3 | 4.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 0.8 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 4.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 2.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 2.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 2.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 2.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 1.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 3.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.7 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.2 | 3.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 4.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 2.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 2.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 4.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 3.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 11.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.9 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 0.6 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 3.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 6.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 4.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 3.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 2.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 4.5 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.2 | 1.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.5 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 2.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 0.5 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.2 | 4.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 7.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 5.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 23.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 7.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 4.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0019966 | C-X-C chemokine binding(GO:0019958) interleukin-1 binding(GO:0019966) tumor necrosis factor binding(GO:0043120) |
| 0.1 | 1.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 1.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 1.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.0 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.9 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 1.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 5.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 14.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 5.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 1.3 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.6 | GO:0032184 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) SUMO polymer binding(GO:0032184) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.6 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.6 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 1.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.7 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 8.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 4.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 1.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 2.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 3.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.5 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 1.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 2.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 4.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 8.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 2.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.6 | GO:0001653 | peptide receptor activity(GO:0001653) G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 2.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 8.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.0 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 6.6 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.7 | 4.4 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.6 | 19.4 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.5 | 15.1 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.5 | 2.0 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 3.9 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 3.8 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 7.9 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.4 | 1.8 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.3 | 5.7 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 3.3 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.3 | 4.1 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 10.6 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.3 | 1.7 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 8.8 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.3 | 9.6 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.3 | 15.7 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.3 | 13.7 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.3 | 9.3 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.3 | 16.8 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.3 | 2.1 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 4.2 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.3 | 3.6 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 4.6 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 4.3 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.2 | 4.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.2 | 12.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 6.4 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.2 | 3.3 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.2 | 4.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 2.7 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.2 | 7.4 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 4.6 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 0.8 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.5 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 8.7 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.5 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.1 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.2 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.8 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.7 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.9 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.4 | PID_ENDOTHELIN_PATHWAY | Endothelins |
| 0.1 | 2.5 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 2.5 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 6.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.7 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 3.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.9 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.7 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.9 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.5 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 0.5 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 2.1 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.1 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.1 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 6.1 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 0.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.4 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.6 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 1.2 | 12.0 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.9 | 1.8 | REACTOME_PD1_SIGNALING | Genes involved in PD-1 signaling |
| 0.9 | 6.2 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.8 | 13.8 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 23.1 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 8.3 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.6 | 13.8 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.6 | 5.2 | REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 2.4 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.5 | 6.2 | REACTOME_ACTIVATED_POINT_MUTANTS_OF_FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.5 | 9.3 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 1.3 | REACTOME_SHC_RELATED_EVENTS | Genes involved in SHC-related events |
| 0.4 | 1.3 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 3.1 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 13.1 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 8.6 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.3 | 3.8 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 3.9 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 5.3 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 5.3 | REACTOME_NRIF_SIGNALS_CELL_DEATH_FROM_THE_NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 3.9 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 10.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 7.3 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.3 | 4.9 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 4.0 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 7.0 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.3 | 3.3 | REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 1.7 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 5.9 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 4.2 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.2 | 2.2 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.7 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 2.1 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 7.4 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 4.6 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.3 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.5 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.2 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 0.5 | REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.2 | 2.5 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 11.4 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.2 | 0.3 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 3.3 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.8 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 1.6 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.7 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 0.5 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 2.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 3.0 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.1 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.9 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.6 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.7 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 3.6 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 9.6 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.7 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.4 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.7 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.3 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.3 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.5 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.1 | 2.2 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.8 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.8 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.5 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.3 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.2 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.5 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.9 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.6 | REACTOME_GAP_JUNCTION_TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 1.0 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.0 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.5 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.4 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.7 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.7 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.3 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.5 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.4 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |