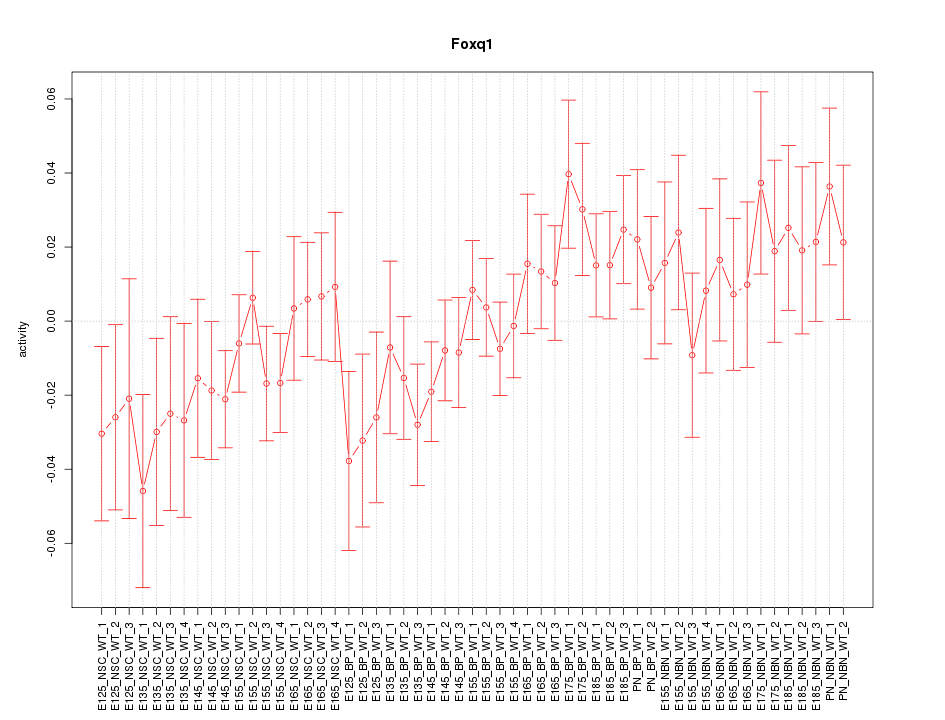
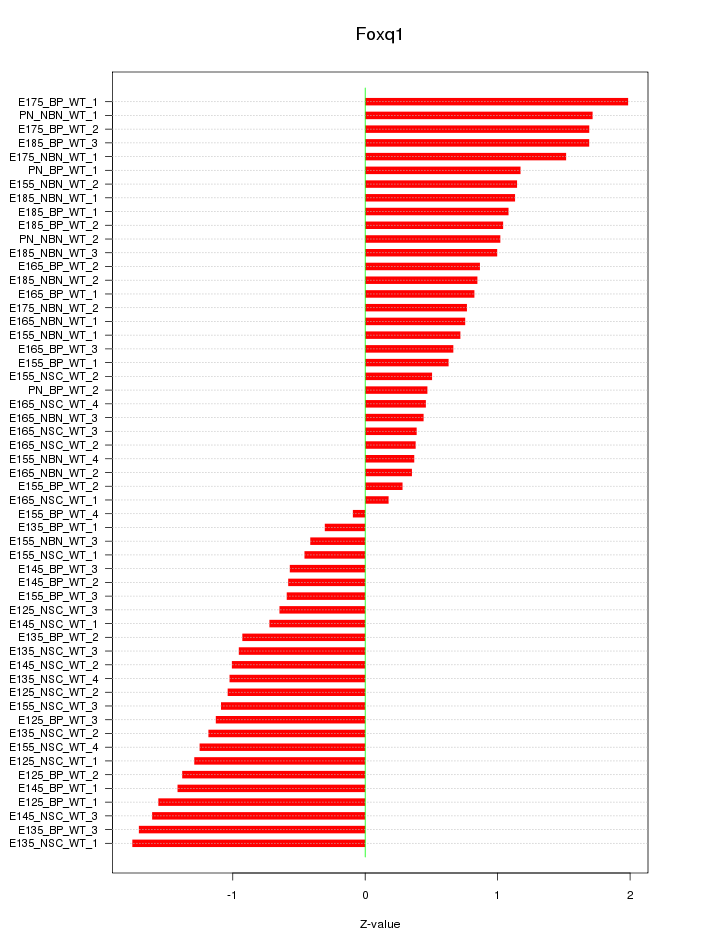
Motif ID: Foxq1
Z-value: 1.034
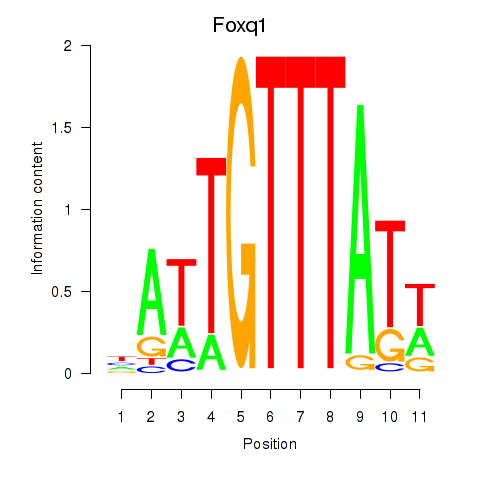
Transcription factors associated with Foxq1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxq1 | ENSMUSG00000038415.8 | Foxq1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxq1 | mm10_v2_chr13_+_31558157_31558176 | 0.25 | 6.3e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 2.9 | 11.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 2.2 | 4.4 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.6 | 4.9 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.6 | 4.8 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) regulation of bleb assembly(GO:1904170) |
| 0.9 | 8.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 | 2.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.6 | 4.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.5 | 2.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.5 | 2.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 8.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 | 3.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.4 | 2.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 1.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.4 | 1.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 3.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 13.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 4.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 3.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.3 | 3.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 7.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 8.0 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.3 | 6.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 2.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 1.2 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 | 1.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 1.2 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.2 | 1.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 2.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.7 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.2 | 2.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 28.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 3.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 1.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.9 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.7 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 3.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 2.9 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 9.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 3.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.1 | 0.8 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 3.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.4 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 9.8 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 1.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 2.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 2.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 2.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 11.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.1 | 10.8 | GO:0000805 | X chromosome(GO:0000805) |
| 0.5 | 2.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 4.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 4.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.3 | 8.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 9.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 2.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 2.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.2 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 8.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 4.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 8.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 4.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 27.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 5.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 3.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 1.5 | 11.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.9 | 3.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.8 | 4.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.8 | 2.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.4 | 2.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 3.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 1.9 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.3 | 2.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 2.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 2.9 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 3.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 2.0 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 8.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 20.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 3.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 5.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 10.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 11.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 6.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.6 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 1.3 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.9 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.4 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 1.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 3.2 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.8 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 3.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.9 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.0 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.7 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.5 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.8 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.0 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.0 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 2.2 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |