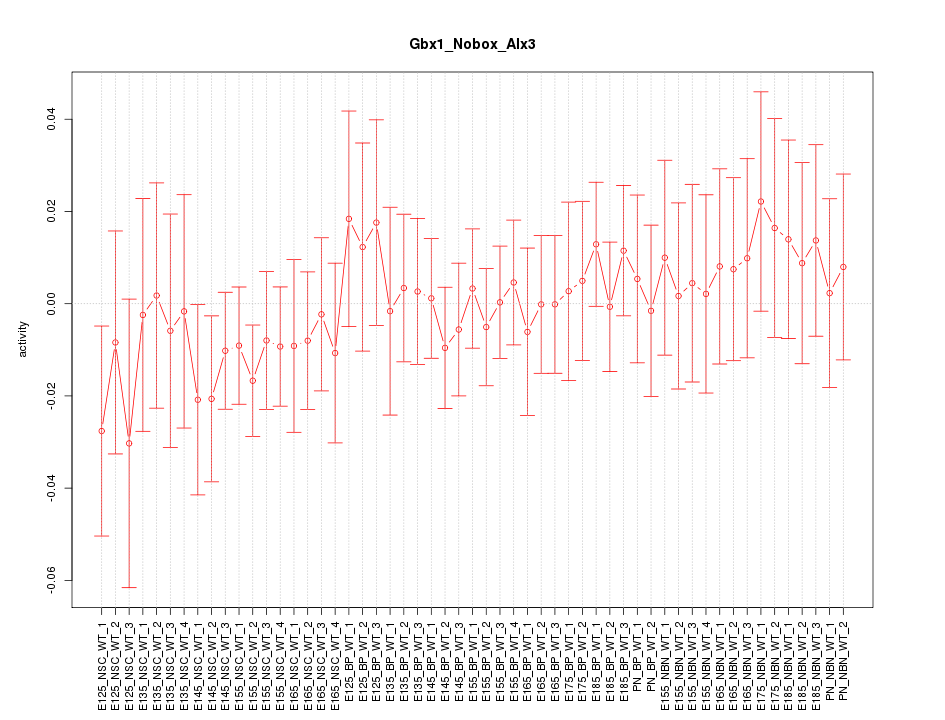
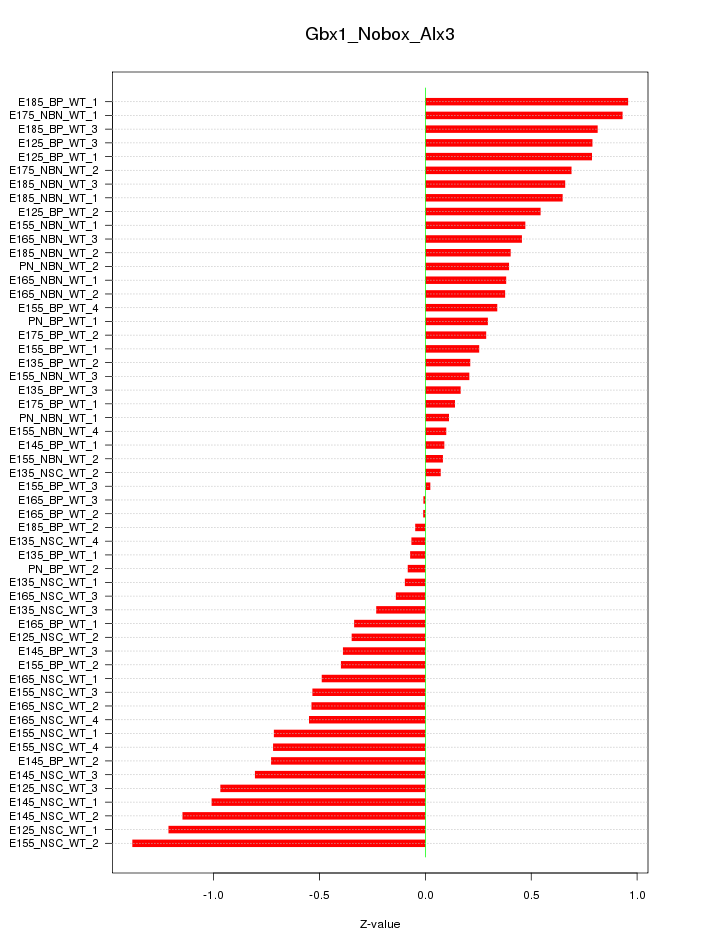
Motif ID: Gbx1_Nobox_Alx3
Z-value: 0.564
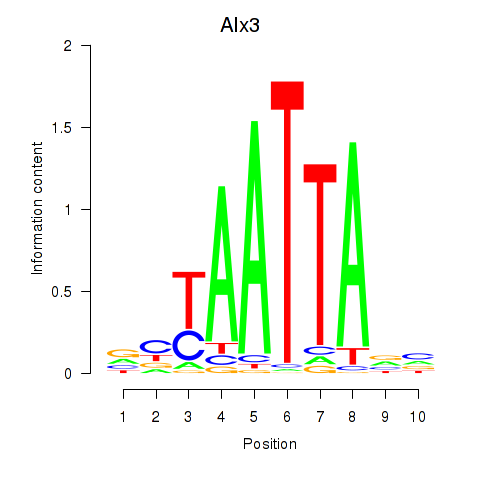
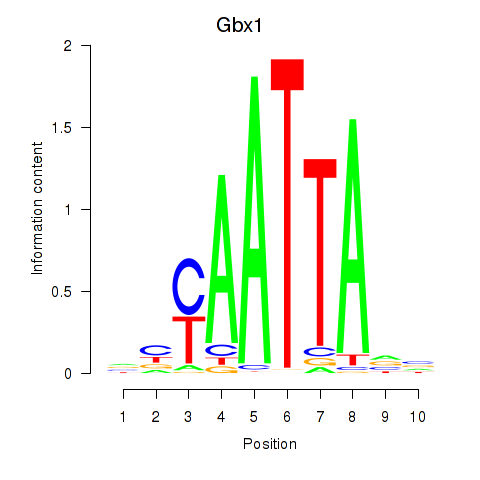
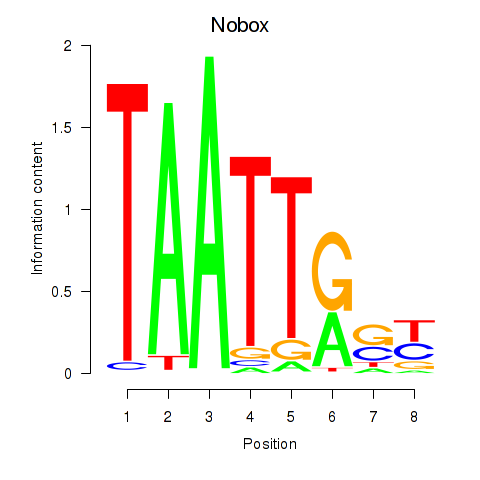
Transcription factors associated with Gbx1_Nobox_Alx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Alx3 | ENSMUSG00000014603.1 | Alx3 |
| Gbx1 | ENSMUSG00000067724.4 | Gbx1 |
| Nobox | ENSMUSG00000029736.9 | Nobox |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Alx3 | mm10_v2_chr3_+_107595031_107595164 | 0.11 | 4.1e-01 | Click! |
| Gbx1 | mm10_v2_chr5_-_24527276_24527276 | -0.03 | 8.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.6 | 1.8 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.6 | 1.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 2.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.5 | 2.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.5 | 1.6 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.5 | 1.5 | GO:0071336 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.5 | 3.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 | 5.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.5 | 2.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 1.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.5 | 6.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 1.3 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 2.5 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 | 3.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 | 3.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 1.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 2.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 0.6 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 | 2.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 0.9 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.3 | 2.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 | 6.8 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.3 | 1.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 1.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 0.8 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.3 | 2.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.7 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.7 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.7 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.2 | 0.7 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 0.9 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.2 | 1.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.4 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.2 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 1.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 2.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.2 | 1.3 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 0.5 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 4.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 1.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 1.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.1 | 0.7 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.4 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.6 | GO:0070627 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) ferrous iron import(GO:0070627) |
| 0.1 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 3.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 3.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.6 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.0 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 2.8 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.3 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.3 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 0.4 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 0.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 3.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 3.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.4 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 5.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 1.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.3 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.2 | GO:0046878 | corticosterone secretion(GO:0035934) positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 1.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.6 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 1.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 1.1 | GO:2000726 | negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 1.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.7 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 1.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 1.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 2.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 3.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 3.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 2.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 2.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.9 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 1.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.9 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.5 | 2.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 2.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.4 | 6.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.3 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 1.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 2.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 5.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.4 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.5 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 2.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.6 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 2.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 5.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 2.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 2.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.9 | 3.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 3.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 4.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.4 | 3.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 1.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 2.4 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.3 | 2.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.7 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 1.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 2.2 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 1.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 3.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 1.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 1.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 1.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 2.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 0.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 2.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 3.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 2.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.6 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 1.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 2.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 10.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 2.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 2.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 4.4 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 6.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.7 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 1.6 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.2 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 0.9 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.1 | 4.2 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.8 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.4 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.1 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 0.2 | REACTOME_SHC1_EVENTS_IN_EGFR_SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 5.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.4 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.3 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.8 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.3 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.7 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.5 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.4 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.6 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.0 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.6 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.3 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.8 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.4 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 5.8 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.0 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.0 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.3 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.1 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.0 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.6 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |