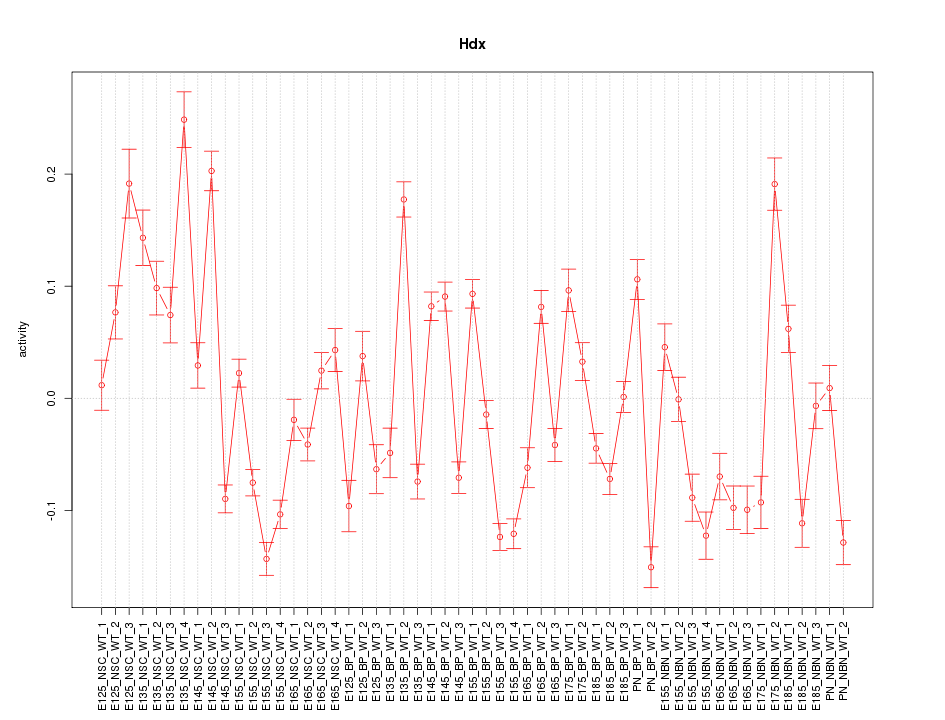
Motif ID: Hdx
Z-value: 5.499
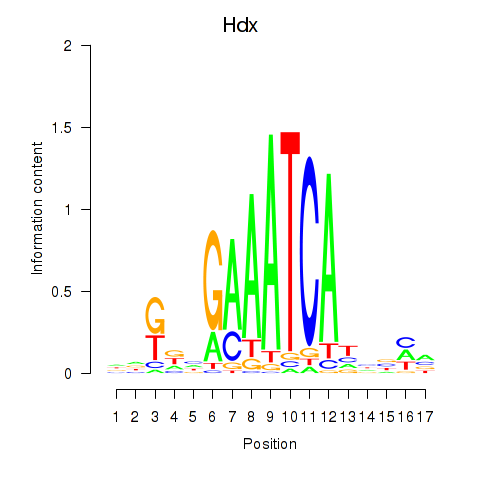
Transcription factors associated with Hdx:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hdx | ENSMUSG00000034551.6 | Hdx |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hdx | mm10_v2_chrX_-_111697069_111697127 | -0.07 | 6.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 24.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 3.8 | 11.5 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 2.8 | 8.5 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 2.1 | 10.7 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 1.3 | 5.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.9 | 4.4 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.6 | 4.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.6 | 1.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.6 | 2.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.6 | 2.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 1.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.5 | 3.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.5 | 1.8 | GO:0001692 | histamine metabolic process(GO:0001692) histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.4 | 8.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 11.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 4.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 11.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 4.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 7.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 3.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 2.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 7.9 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 3.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1936.1 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 8.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.3 | 7.9 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.8 | 3.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.7 | 11.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.4 | 1.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 11.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 3.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 4.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 4.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 3.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2004.2 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.5 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 1.7 | 5.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.6 | 11.5 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 1.3 | 8.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.1 | 4.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.7 | 10.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.5 | 9.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 3.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 4.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 2.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 5.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 11.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 3.0 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 4.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 4.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 26.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1975.9 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.4 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.3 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.3 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.7 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 5.1 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 11.6 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 2.3 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 4.4 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 4.1 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 2.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.2 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.7 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.1 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.8 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |