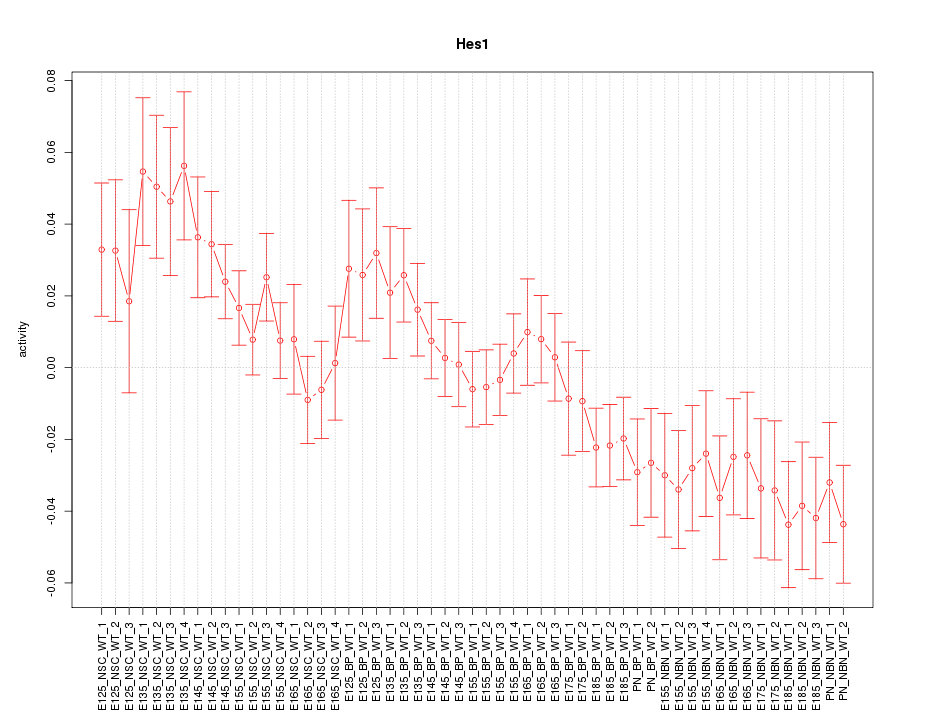
Motif ID: Hes1
Z-value: 1.628
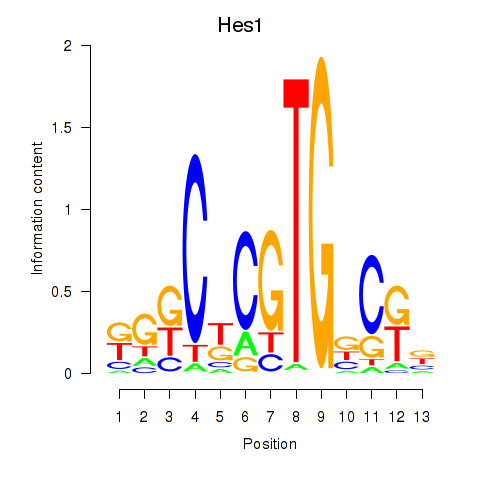
Transcription factors associated with Hes1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hes1 | ENSMUSG00000022528.7 | Hes1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes1 | mm10_v2_chr16_+_30065333_30065351 | 0.63 | 2.3e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 119.9 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 8.0 | 23.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 5.6 | 22.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 3.8 | 15.3 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 3.8 | 15.0 | GO:0003360 | brainstem development(GO:0003360) |
| 3.7 | 14.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 3.0 | 12.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 2.6 | 7.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 2.4 | 7.3 | GO:0003195 | tricuspid valve formation(GO:0003195) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 2.4 | 9.7 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 2.4 | 7.1 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.8 | 7.1 | GO:0060032 | notochord regression(GO:0060032) |
| 1.6 | 1.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 1.6 | 6.5 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.3 | 5.3 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 1.3 | 11.5 | GO:0001842 | neural fold formation(GO:0001842) |
| 1.2 | 8.1 | GO:0046549 | phenol-containing compound catabolic process(GO:0019336) retinal cone cell development(GO:0046549) |
| 1.1 | 3.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 1.1 | 4.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.1 | 6.5 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 1.0 | 3.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 1.0 | 4.9 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.9 | 7.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.9 | 5.7 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) macropinocytosis(GO:0044351) |
| 0.9 | 4.6 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.8 | 2.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.8 | 5.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.8 | 2.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.8 | 2.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.8 | 5.5 | GO:0089700 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) |
| 0.7 | 2.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.7 | 3.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.7 | 10.0 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.7 | 2.8 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.7 | 6.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 7.2 | GO:0030432 | peristalsis(GO:0030432) |
| 0.6 | 3.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.6 | 1.7 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.5 | 2.7 | GO:0048133 | NK T cell differentiation(GO:0001865) germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.5 | 3.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 | 1.5 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.5 | 4.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.5 | 3.6 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.4 | 9.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.4 | 5.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 1.3 | GO:0072554 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.4 | 2.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 1.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.4 | 1.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.4 | 2.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.4 | 2.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 2.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.4 | 1.9 | GO:1903598 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.4 | 1.9 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.4 | 1.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 2.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.3 | 1.0 | GO:0033122 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 2.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 1.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.3 | 5.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 0.8 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.3 | 6.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 0.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 11.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.3 | 2.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 2.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.0 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.2 | 2.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 8.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.2 | 0.9 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.2 | 2.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 1.6 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 | 3.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 5.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 2.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.6 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 1.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.2 | 0.6 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.2 | 4.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.2 | 3.8 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.2 | 5.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 3.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.2 | 0.7 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 1.7 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.2 | 0.5 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 8.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 1.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 4.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 4.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 0.8 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 1.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 8.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 5.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.4 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.3 | GO:0010455 | positive regulation of cell fate commitment(GO:0010455) regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 2.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 2.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 2.9 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.1 | 8.8 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.1 | 2.0 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 2.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 14.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 3.4 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 2.4 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 2.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.1 | 0.7 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 3.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.4 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 3.3 | GO:0009306 | protein secretion(GO:0009306) |
| 0.1 | 1.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.5 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.8 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 0.9 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.2 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 0.6 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.1 | 6.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.2 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.1 | 13.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 0.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 1.8 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 1.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 1.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.3 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 2.9 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.0 | 1.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 3.1 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 5.8 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 1.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.5 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 2.4 | 9.6 | GO:0090537 | CERF complex(GO:0090537) |
| 1.6 | 9.4 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.9 | 2.8 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.9 | 4.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.9 | 11.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 12.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.7 | 4.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.7 | 6.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.6 | 2.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 7.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.5 | 7.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.5 | 6.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 8.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 6.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.4 | 1.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.4 | 1.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 3.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 1.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 0.8 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 4.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 2.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 10.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 5.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 4.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 9.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 1.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 5.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 3.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 24.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 1.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.5 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 2.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 1.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 3.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.9 | GO:0044754 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 8.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 7.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 6.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 21.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 12.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 4.6 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 29.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 3.6 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.3 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 4.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 3.4 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 22.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 3.1 | 120.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 2.6 | 7.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 2.5 | 15.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 2.4 | 7.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 2.4 | 7.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 2.2 | 6.5 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.8 | 7.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 1.4 | 2.9 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.1 | 6.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 3.0 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.9 | 5.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.9 | 3.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.8 | 6.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.7 | 5.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.7 | 3.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 4.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.7 | 2.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.7 | 2.8 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.6 | 4.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 3.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.5 | 2.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.5 | 4.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.5 | 1.9 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.4 | 6.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 25.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 13.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 4.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.4 | 1.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 4.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 1.9 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.4 | 3.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 11.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 2.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.3 | 5.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 0.9 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.3 | 12.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 9.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.3 | 3.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 0.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 10.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 2.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 4.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 6.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 7.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 1.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 5.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 1.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 6.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 4.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 1.1 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.2 | 3.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.2 | 12.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.2 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 3.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 9.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 12.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.9 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 2.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 2.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 14.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 0.8 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 2.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 18.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 4.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 3.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 2.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 2.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 2.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 10.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 2.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 2.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 130.3 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.4 | 23.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.3 | 7.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 7.7 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.3 | 8.2 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 3.6 | PID_ERBB1_RECEPTOR_PROXIMAL_PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.2 | 12.0 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 7.4 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.2 | 15.2 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.2 | 8.1 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.2 | 4.9 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 22.0 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.7 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.2 | 2.8 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.2 | 1.6 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 1.7 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.2 | 8.6 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 10.8 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 7.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.5 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.2 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.0 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 6.3 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 4.1 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.1 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.5 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.0 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 2.9 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.4 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.0 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 119.9 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.8 | 13.5 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 8.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 11.7 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.5 | 3.7 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 6.8 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 7.6 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.4 | 12.1 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.4 | 3.2 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 3.1 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 11.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 3.4 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.3 | 23.0 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 5.2 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.3 | 42.8 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.3 | 2.5 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 3.2 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 4.3 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 7.5 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.2 | 3.5 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.9 | REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 4.9 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 6.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 1.2 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 6.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 7.5 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.9 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.2 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 5.1 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.3 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 9.2 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.5 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.0 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.2 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.8 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.9 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.7 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 1.9 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 4.8 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.1 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.3 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |