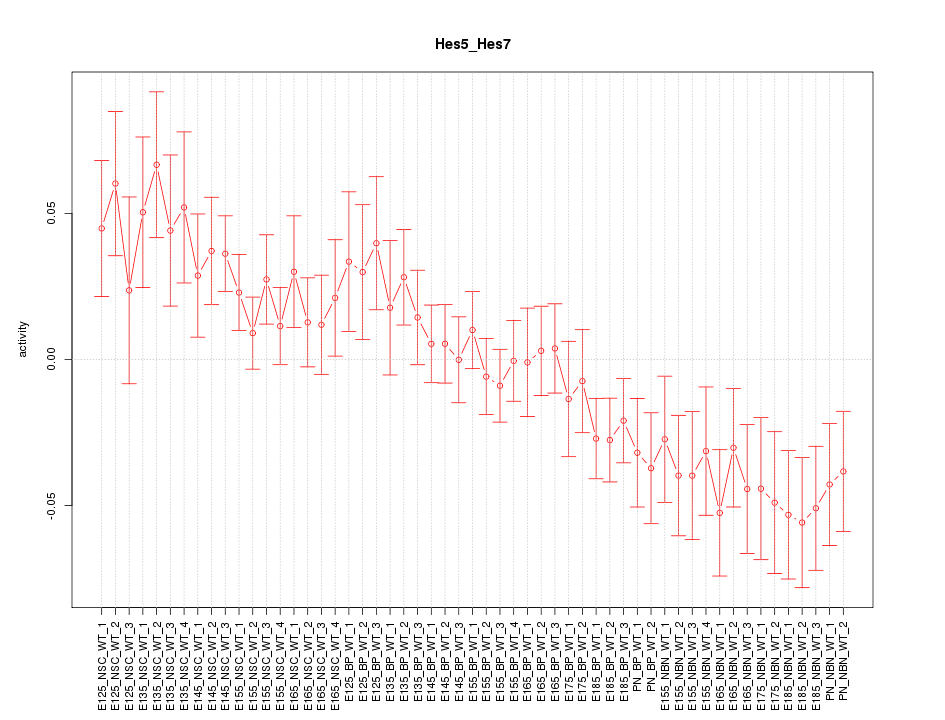
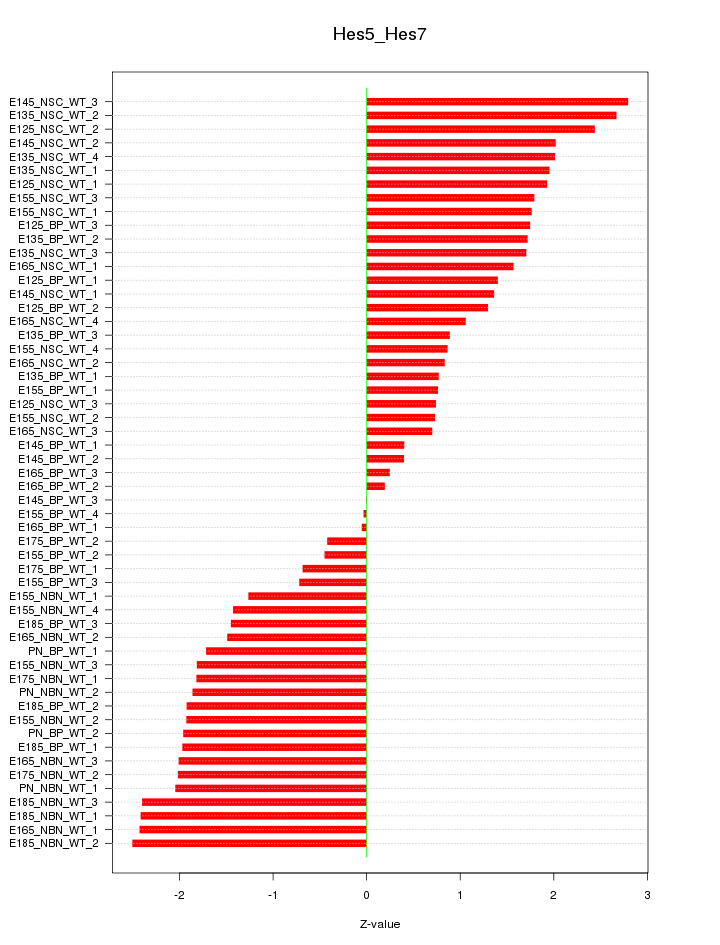
Motif ID: Hes5_Hes7
Z-value: 1.595
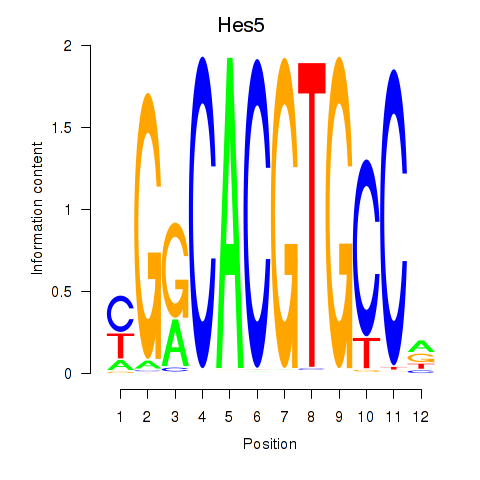
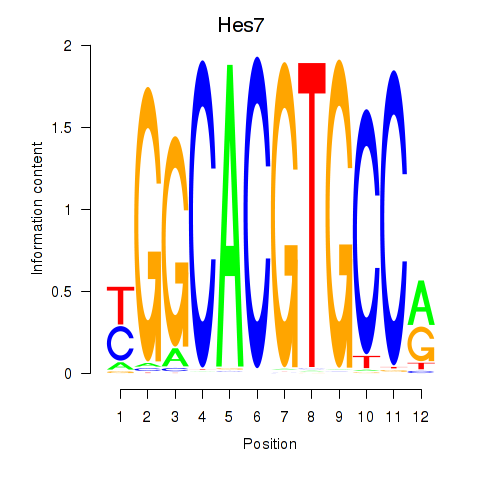
Transcription factors associated with Hes5_Hes7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hes5 | ENSMUSG00000048001.7 | Hes5 |
| Hes7 | ENSMUSG00000023781.2 | Hes7 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes5 | mm10_v2_chr4_+_154960915_154960930 | 0.70 | 2.6e-09 | Click! |
| Hes7 | mm10_v2_chr11_+_69120404_69120404 | -0.05 | 7.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.5 | 193.2 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 6.7 | 20.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 2.7 | 8.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 2.3 | 22.9 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 1.8 | 5.5 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.5 | 4.6 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.3 | 3.8 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 1.2 | 8.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.2 | 9.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 1.1 | 3.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.0 | 9.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.8 | 3.2 | GO:0015744 | succinate transport(GO:0015744) |
| 0.7 | 5.7 | GO:0090091 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.6 | 3.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.4 | 4.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 7.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.3 | 1.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.3 | 1.6 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 2.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.3 | 5.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 2.5 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.3 | 2.0 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 7.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 2.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 26.2 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.2 | 21.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.2 | 0.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.2 | 1.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 8.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 1.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 2.7 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.6 | GO:0034143 | regulation of toll-like receptor 4 signaling pathway(GO:0034143) |
| 0.0 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.5 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 1.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 6.4 | GO:1901657 | glycosyl compound metabolic process(GO:1901657) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.7 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 4.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 6.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 3.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 20.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 9.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 8.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 7.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 7.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 150.5 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 7.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 2.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 6.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 3.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 20.2 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 37.3 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 2.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.3 | GO:1990234 | transferase complex(GO:1990234) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 193.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 3.1 | 9.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.9 | 5.7 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.9 | 7.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 1.4 | 5.5 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 1.1 | 3.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 1.1 | 8.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.8 | 3.8 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.7 | 5.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.7 | 8.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.5 | 4.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 20.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.2 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.3 | 2.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 9.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 2.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 4.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 14.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 17.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 2.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 3.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 14.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 7.7 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 4.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 193.2 | PID_IGF1_PATHWAY | IGF1 pathway |
| 1.0 | 9.3 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.7 | 22.9 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 15.7 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 20.0 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 3.2 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 4.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 3.1 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 8.1 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 5.9 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.7 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.2 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID_BMP_PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 193.2 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.9 | 9.3 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 5.9 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.8 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.7 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.5 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 5.7 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 7.6 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 26.7 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.2 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 4.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 9.5 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.4 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.6 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |