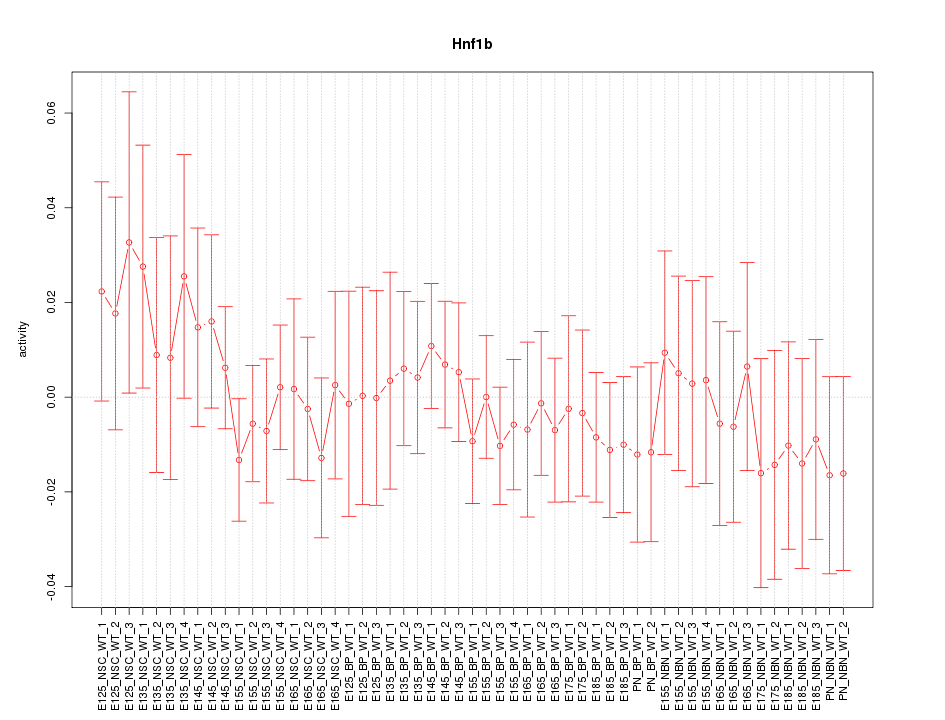
Motif ID: Hnf1b
Z-value: 0.560
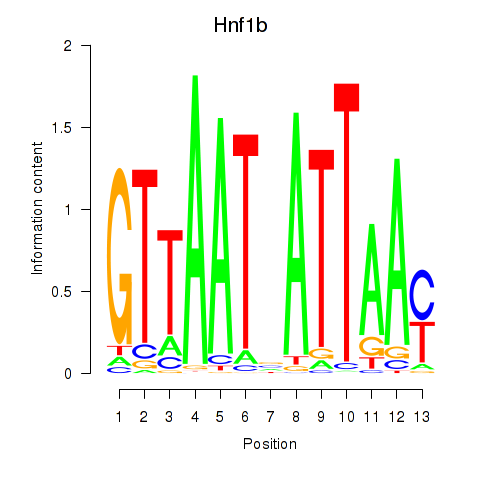
Transcription factors associated with Hnf1b:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hnf1b | ENSMUSG00000020679.5 | Hnf1b |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.8 | 2.5 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 0.7 | 2.8 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.6 | 2.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) negative regulation of low-density lipoprotein particle clearance(GO:0010989) positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 0.6 | 1.8 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.5 | 1.4 | GO:0009814 | defense response, incompatible interaction(GO:0009814) |
| 0.4 | 1.7 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.4 | 1.9 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.4 | 1.9 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.3 | 1.6 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.3 | 1.5 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 1.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.6 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.2 | 0.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.6 | GO:0050787 | glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 1.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 0.5 | GO:1904956 | regulation of endodermal cell fate specification(GO:0042663) Wnt signaling pathway involved in somitogenesis(GO:0090244) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.6 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.6 | GO:0032275 | luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.1 | 0.6 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.1 | 2.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 2.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.1 | 3.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.9 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 2.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.8 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.3 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 2.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 3.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 0.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.4 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0035482 | gastric motility(GO:0035482) gastric emptying(GO:0035483) negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 1.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.5 | 1.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 1.7 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.4 | 2.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 0.9 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 0.6 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 3.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 2.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.6 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 2.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 5.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.8 | 2.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.8 | 2.5 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.4 | 1.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 1.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.9 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.2 | 0.9 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.2 | 0.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.2 | 1.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.6 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.2 | 5.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.9 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 2.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 2.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 3.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 1.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 0.9 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 3.6 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.7 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.2 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.9 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.9 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.8 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.9 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.8 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.2 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.5 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.6 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.3 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.5 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |