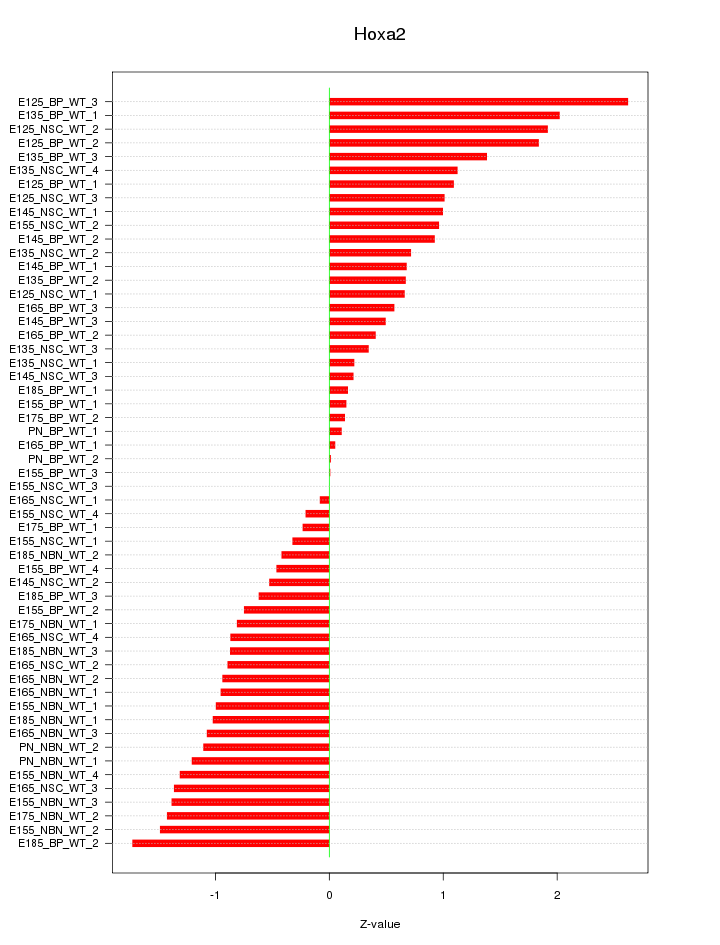
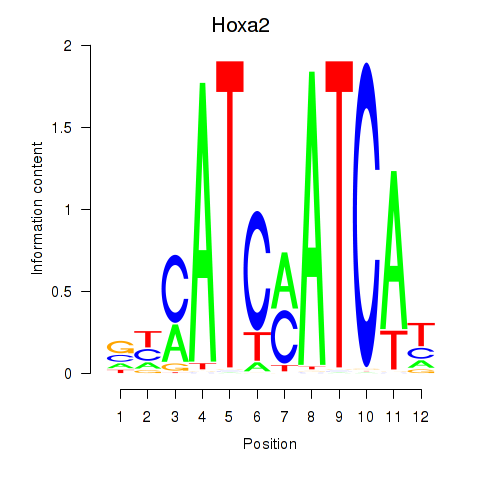
Motif ID: Hoxa2
Z-value: 0.994
Transcription factors associated with Hoxa2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa2 | ENSMUSG00000014704.8 | Hoxa2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.3 | 17.2 | GO:0060032 | notochord regression(GO:0060032) |
| 2.8 | 24.8 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 2.1 | 10.7 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 2.1 | 14.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 1.9 | 9.7 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.4 | 5.5 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 1.1 | 3.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.0 | 3.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.0 | 3.0 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.8 | 3.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.7 | 2.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.7 | 1.4 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.6 | 1.8 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.6 | 3.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.6 | 5.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.6 | 1.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.6 | 3.4 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.6 | 3.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.6 | 2.8 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.5 | 2.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 1.0 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.5 | 2.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 1.5 | GO:1900041 | intestinal D-glucose absorption(GO:0001951) negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.5 | 1.9 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.5 | 1.9 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.5 | 1.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.4 | 1.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.4 | 1.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.4 | 1.2 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 4.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.4 | 1.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 3.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 3.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.3 | 3.6 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.3 | 1.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 0.6 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.3 | 1.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.3 | 1.2 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.3 | 1.7 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 1.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 2.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.3 | 0.8 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 1.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.3 | 0.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 5.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.3 | 0.8 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 1.7 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.2 | 2.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 0.7 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 1.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 0.2 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.2 | 0.6 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.2 | 0.8 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.2 | 1.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 1.0 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 4.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 2.8 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 4.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 14.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.2 | 1.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 0.5 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.2 | 0.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 0.7 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 1.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 2.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.4 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 1.0 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 15.6 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.1 | 1.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 4.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.2 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.9 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 2.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.8 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 1.5 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.1 | 0.5 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.7 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.4 | GO:0003211 | cardiac ventricle formation(GO:0003211) |
| 0.1 | 0.4 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.2 | GO:2000224 | testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 1.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.9 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.7 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 4.1 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.8 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 4.8 | GO:0007051 | spindle organization(GO:0007051) |
| 0.1 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.2 | GO:0061156 | pulmonary artery morphogenesis(GO:0061156) |
| 0.1 | 0.3 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 3.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.0 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.1 | 0.3 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 1.2 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 0.9 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.6 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 5.3 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.4 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 | 1.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:1902713 | regulation of interferon-gamma secretion(GO:1902713) |
| 0.1 | 0.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 1.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 3.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.7 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 1.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 2.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 2.5 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 1.8 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:2001184 | regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 1.5 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 1.8 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.6 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 1.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 2.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.1 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 1.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 2.1 | GO:0043487 | regulation of RNA stability(GO:0043487) |
| 0.0 | 1.0 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.4 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.4 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.7 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 5.0 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 20.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.3 | 17.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.7 | 2.7 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.7 | 2.0 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 3.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.5 | 2.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 4.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 4.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 1.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 2.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 3.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 1.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 3.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.2 | 1.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 2.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 2.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 2.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 1.0 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 11.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.5 | GO:0033093 | multivesicular body membrane(GO:0032585) Weibel-Palade body(GO:0033093) |
| 0.1 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 5.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 7.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 1.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 4.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 3.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 4.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.7 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 4.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 5.4 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 1.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 1.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 27.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.9 | 5.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.2 | 9.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.0 | 3.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.8 | 15.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.7 | 2.2 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.7 | 3.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 2.4 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.6 | 2.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.5 | 2.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.5 | 2.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 4.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 17.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.5 | 3.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.5 | 1.4 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 1.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 1.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.4 | 2.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 3.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 1.0 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 10.7 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.3 | 1.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.3 | 14.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 1.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.2 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 0.8 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.3 | 0.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.2 | 0.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 1.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 0.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.7 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 5.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 0.8 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 2.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 2.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 0.5 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.2 | 0.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 1.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 3.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 0.6 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.7 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.4 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 6.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 3.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.8 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 5.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.0 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 2.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 3.1 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.1 | 1.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 3.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.9 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 6.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 2.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 2.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 12.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 24.3 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.5 | 17.2 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 7.9 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 8.8 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 2.9 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 16.5 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.0 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 4.6 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 2.7 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.3 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.0 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.1 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.0 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.2 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.7 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.8 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.1 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.7 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 15.4 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 25.0 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.5 | 9.3 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 3.3 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 24.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 3.1 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 2.2 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 4.1 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.2 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 4.9 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.5 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.0 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 5.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.7 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.2 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.0 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 5.2 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.9 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.0 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 2.4 | REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 7.0 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 3.6 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 4.4 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.3 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 5.3 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.5 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.4 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |