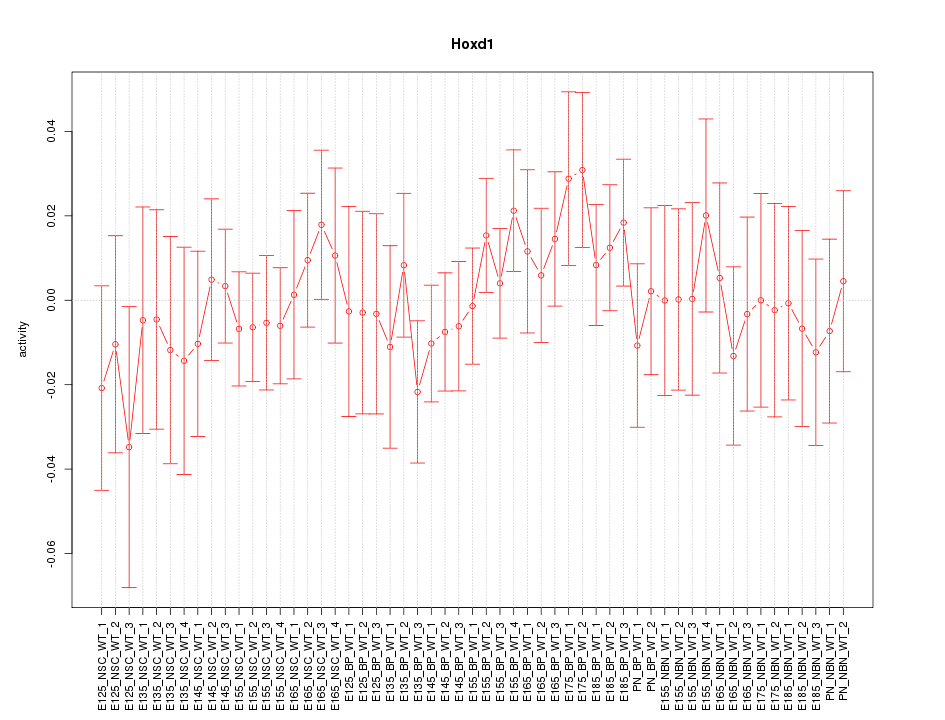
Motif ID: Hoxd1
Z-value: 0.644
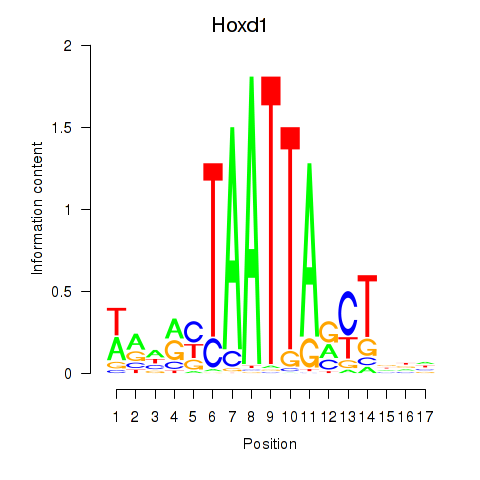
Transcription factors associated with Hoxd1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxd1 | ENSMUSG00000042448.4 | Hoxd1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.6 | 3.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.6 | 1.9 | GO:2000564 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.6 | 1.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.4 | 1.5 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 | 1.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.3 | 1.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 1.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 3.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 1.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 0.9 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 2.7 | GO:1902018 | regulation of mitotic spindle assembly(GO:1901673) negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 1.2 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 0.6 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.6 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.4 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:2000338 | positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.6 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.6 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 1.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.8 | GO:0036159 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.1 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.4 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 0.1 | 1.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.5 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.4 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 0.5 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 1.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0098835 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 0.3 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 3.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.9 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 1.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 3.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.7 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 1.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.5 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 2.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.1 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 2.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0005402 | cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0035615 | clathrin heavy chain binding(GO:0032050) clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 3.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 3.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.0 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.9 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 1.6 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 1.1 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID_IL1_PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.7 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 2.1 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 4.1 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.2 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |