Motif ID: Irx6_Irx2_Irx3
Z-value: 0.749
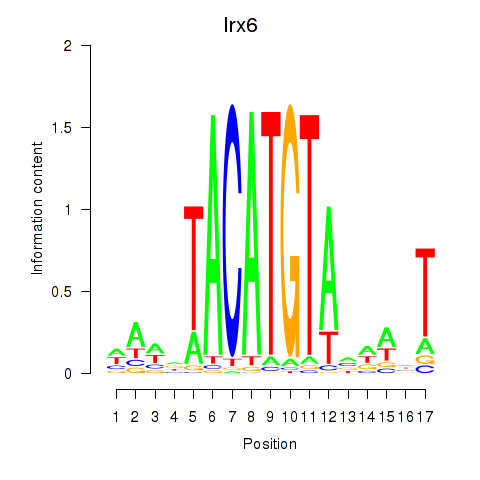
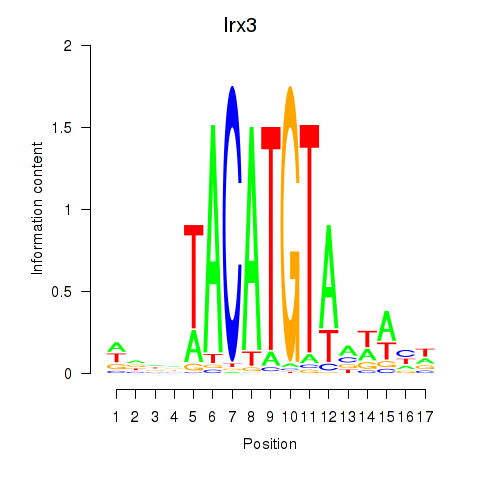
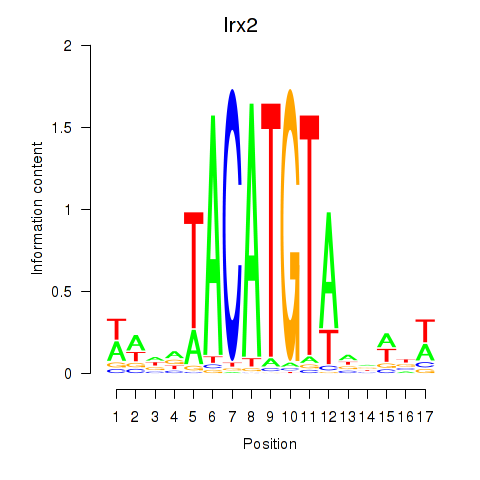
Transcription factors associated with Irx6_Irx2_Irx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Irx2 | ENSMUSG00000001504.9 | Irx2 |
| Irx3 | ENSMUSG00000031734.11 | Irx3 |
| Irx6 | ENSMUSG00000031738.8 | Irx6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irx2 | mm10_v2_chr13_+_72632597_72632625 | -0.23 | 8.9e-02 | Click! |
| Irx3 | mm10_v2_chr8_-_91801547_91801560 | -0.02 | 8.8e-01 | Click! |
| Irx6 | mm10_v2_chr8_+_92674289_92674289 | 0.01 | 9.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 9.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.9 | 3.7 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 2.4 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.4 | 2.1 | GO:0072008 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.4 | 1.6 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 | 1.6 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 | 1.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.4 | 1.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 1.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.3 | 1.5 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 2.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 2.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.6 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.2 | 0.5 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 0.5 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 0.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.5 | GO:2001180 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.2 | 1.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.4 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.1 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.6 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.1 | 0.8 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.1 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.3 | GO:0006507 | GPI anchor release(GO:0006507) |
| 0.1 | 0.4 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.3 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 1.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.3 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 1.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 1.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.2 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 3.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.3 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.3 | GO:0099566 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.3 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 | 0.2 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.4 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.4 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.6 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.6 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.9 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 1.2 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 2.2 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.4 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 1.7 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 1.5 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.5 | 1.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 1.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 0.8 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 1.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.5 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 9.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.6 | 1.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 2.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.4 | 1.1 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.3 | 1.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.6 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.2 | 0.8 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.2 | 1.6 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.2 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 1.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 2.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.4 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.1 | 0.5 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.7 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.5 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.3 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.1 | 0.3 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.3 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 3.8 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.8 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.5 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 1.1 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.8 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.6 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.7 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 1.5 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 2.1 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.2 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 10.1 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.2 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.5 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.3 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.1 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.5 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.2 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.9 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 2.4 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 3.7 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME_SCFSKP2_MEDIATED_DEGRADATION_OF_P27_P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.2 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |





