Motif ID: Isl1
Z-value: 0.369
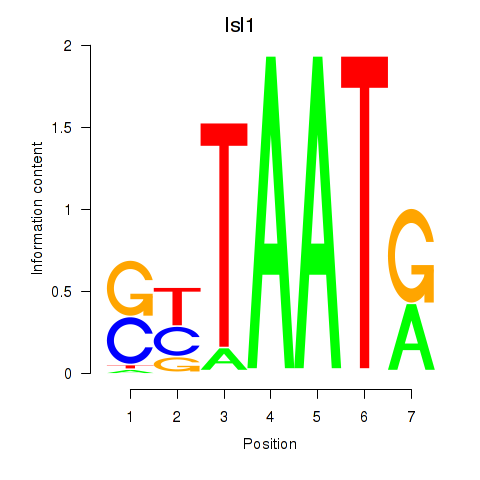
Transcription factors associated with Isl1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Isl1 | ENSMUSG00000042258.7 | Isl1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Isl1 | mm10_v2_chr13_-_116309639_116309699 | 0.06 | 6.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.4 | 3.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 2.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.3 | 1.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 0.7 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 1.4 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.6 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 2.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 1.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.8 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.6 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 1.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 7.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 0.6 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 1.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.7 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID_AP1_PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 0.9 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 2.7 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 0.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |





