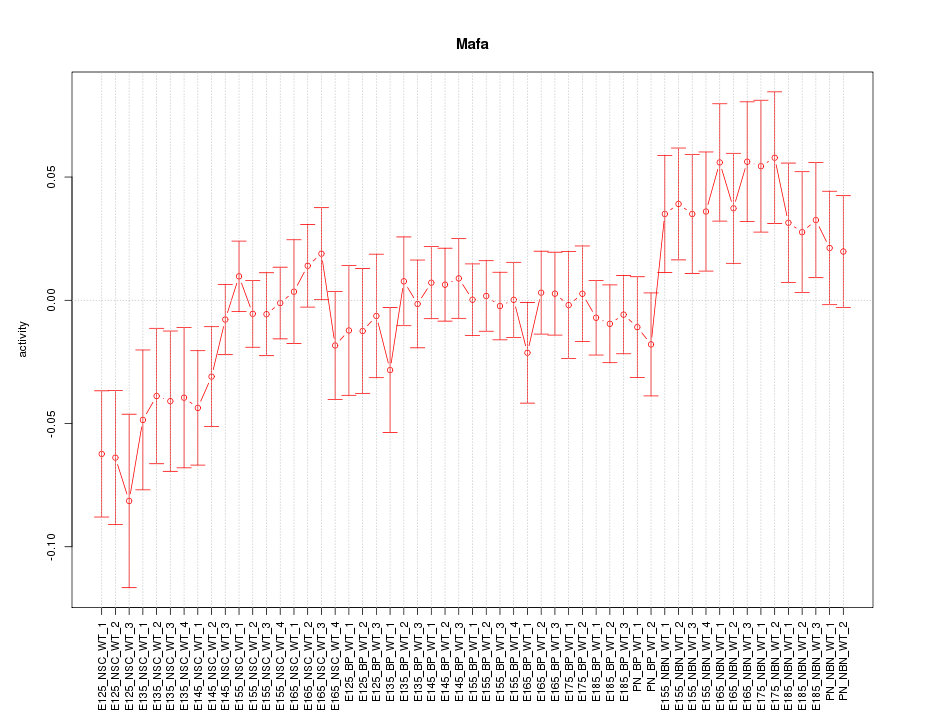
Motif ID: Mafa
Z-value: 1.200
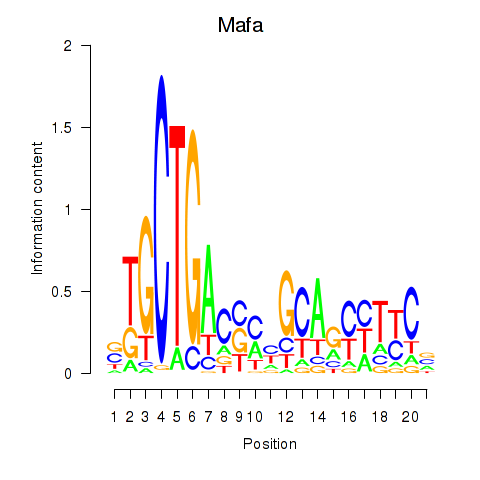
Transcription factors associated with Mafa:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mafa | ENSMUSG00000047591.4 | Mafa |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafa | mm10_v2_chr15_-_75747922_75747922 | -0.44 | 7.9e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 2.6 | 15.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 2.0 | 10.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 1.8 | 22.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.0 | 2.9 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.8 | 3.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.8 | 3.2 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.7 | 13.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 6.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.6 | 3.6 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.6 | 4.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 7.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 4.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.5 | 37.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 4.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.4 | 12.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.3 | 4.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 13.8 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.2 | 0.8 | GO:0048133 | NK T cell differentiation(GO:0001865) germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 2.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.3 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 1.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.4 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 2.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.3 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 7.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 2.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 5.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.1 | 1.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 5.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 4.5 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.1 | 4.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 1.5 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 1.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 2.1 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.1 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.0 | 8.9 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 2.2 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 2.3 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 22.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.8 | 10.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.6 | 5.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.5 | 6.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 13.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 10.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.9 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 2.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 37.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 2.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 42.7 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 4.5 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 3.3 | 10.0 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 1.4 | 4.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.1 | 10.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.1 | 1.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 1.1 | 6.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.7 | 22.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.6 | 3.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 3.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.5 | 2.8 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 1.8 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.4 | 1.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.4 | 14.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 2.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 7.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 2.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 12.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 3.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 5.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 3.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 25.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 5.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 6.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 2.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 4.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.3 | 9.5 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 2.9 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 5.7 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.2 | 6.4 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.5 | 23.6 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 15.4 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 10.3 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 2.2 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 4.3 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 14.4 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 5.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.2 | 6.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.5 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.1 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.9 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.6 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.9 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.1 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |