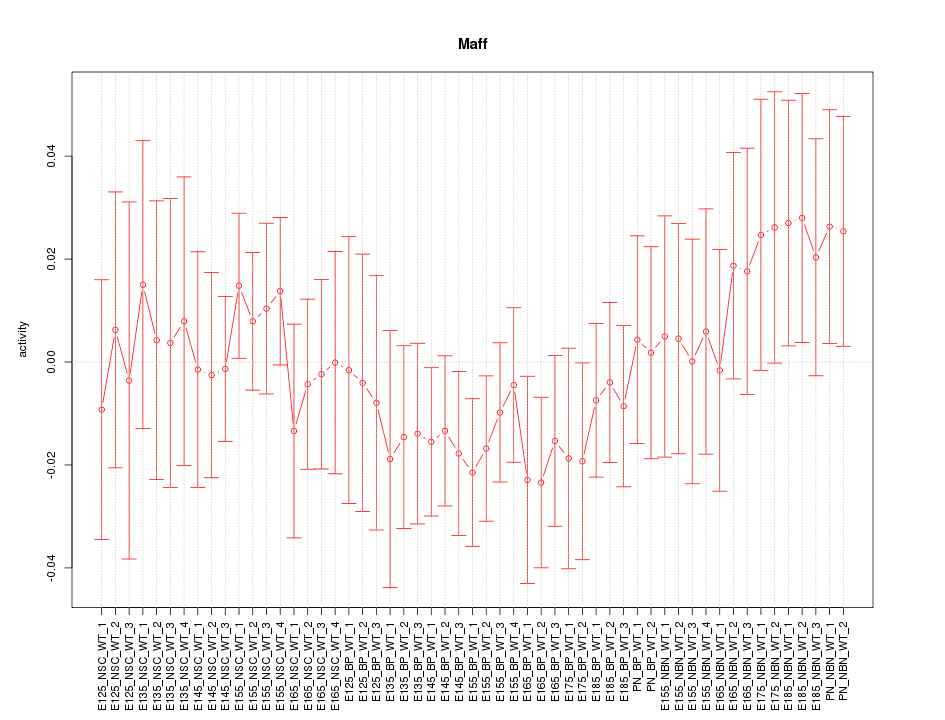
Motif ID: Maff
Z-value: 0.730
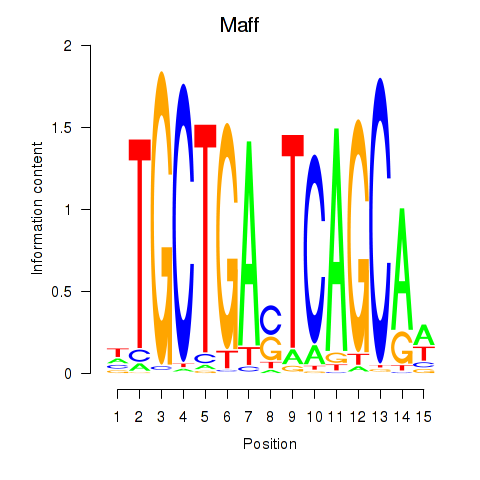
Transcription factors associated with Maff:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Maff | ENSMUSG00000042622.7 | Maff |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Maff | mm10_v2_chr15_+_79347534_79347556 | 0.39 | 3.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0002725 | negative regulation of antigen processing and presentation(GO:0002578) negative regulation of T cell cytokine production(GO:0002725) |
| 1.0 | 4.9 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.7 | 2.2 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.7 | 2.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 | 1.8 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.6 | 5.6 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.5 | 2.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.5 | 2.1 | GO:0035937 | androgen catabolic process(GO:0006710) estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.5 | 2.3 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.5 | 1.4 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 1.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.4 | 1.5 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.3 | 2.4 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.3 | 6.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 0.8 | GO:1900133 | renin secretion into blood stream(GO:0002001) regulation of renin secretion into blood stream(GO:1900133) |
| 0.3 | 1.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 0.8 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 1.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.2 | 1.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 | 0.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 5.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.7 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.8 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.1 | 1.0 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 1.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 2.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 4.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 1.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.9 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.6 | 1.7 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.4 | 1.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.7 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 6.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 4.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 4.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 2.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 3.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 0.9 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.3 | 5.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 1.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 0.8 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.2 | 2.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 2.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.5 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.1 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.6 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 6.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.2 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.2 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 5.6 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.1 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 2.0 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 4.1 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.7 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 2.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.8 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.1 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 1.7 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.8 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.1 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |